
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,525,760 – 12,525,854 |
| Length | 94 |
| Max. P | 0.556982 |

| Location | 12,525,760 – 12,525,854 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 73.61 |
| Shannon entropy | 0.52901 |
| G+C content | 0.47586 |
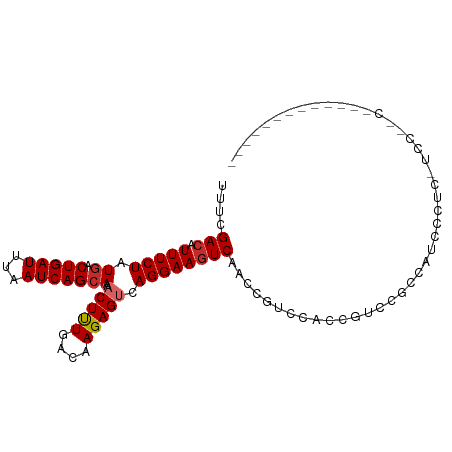
| Mean single sequence MFE | -15.40 |
| Consensus MFE | -10.58 |
| Energy contribution | -10.92 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.49 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.556982 |
| Prediction | RNA |
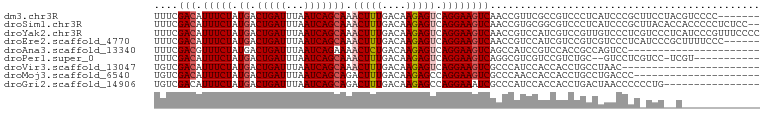
Download alignment: ClustalW | MAF
>dm3.chr3R 12525760 94 + 27905053 UUUCGACAUUUCUAUGACUGAUUUAAUCAGCAAACUUUGACAAGAGUCAGGAAGUCAACCGUUCGCCGUCCCUCAUCCCGCUUCCUACGUCCCC------- ....(((.......((.(((((...)))))))......(((....)))(((((((........................)))))))..)))...------- ( -15.16, z-score = -0.43, R) >droSim1.chr3R 18572927 99 - 27517382 UUUCGACAUUUCUAUGACUGAUUUAAUCAGCAAACUUUGACAAGAGUCAGGAAGUCAACCGUGCGGCGUCCCUCAUCCCGCUUACACCACCCCCUCUCC-- ....(((.(((((.((.(((((...)))))))......(((....)))))))))))......((((.(........)))))..................-- ( -18.30, z-score = -0.94, R) >droYak2.chr3R 16968000 101 - 28832112 UUUCGACAUUUCUAUGACUGAUUUAAUCAGCAAACUUUGACAAGAGUCAGGAAGUCAACCGUCCAUCGUCCGUUGUCCCUCGUCCCUCAUCCCGUUUCCCC ....(((((((((.((.(((((...))))))).....((((....))))))))))((((.(.(....).).))))......)))................. ( -14.70, z-score = 0.20, R) >droEre2.scaffold_4770 8641230 95 - 17746568 UUUCGACAUUUCUAUGACUGAUUUAAUCAGCAAACUUUGACAAGAGUCAGGAAGUCAACCGUCCAUCGUCCGUCGUCCCUCAUCCCGCUUUUCCC------ ...((((((((((.((.(((((...))))))).....((((....))))))))))....((.....))...))))....................------ ( -14.40, z-score = 0.06, R) >droAna3.scaffold_13340 16338672 79 - 23697760 UUUCGACGUUUCUAUGACUGAUUUAAUCAGAAAACUCUGACAAGAGUCAGGAAGUCAGCCAUCCGUCCACCGCCAGUCC---------------------- ....((((..(...((((((((((..(((((....)))))...)))))....)))))...)..))))............---------------------- ( -16.60, z-score = -0.98, R) >droPer1.super_0 11357930 87 + 11822988 UUUCGACAUUUCUAUGACUGAUUUAAUCAGCAAACUUUGACAAGAGUCAGGAAGUCAGGCGUCGUCCGUCUGC--GUCCUCGUCC-UCGU----------- ....(((.......((.(((((...))))))).....((((....))))(((.(.((((((.....)))))))--.)))..))).-....----------- ( -21.20, z-score = -0.60, R) >droVir3.scaffold_13047 13488170 78 - 19223366 UGUCGACAUUUCUAUGACUGAUUUAAUCAGCAAACUUUGACAAGAGUCAGGAAGUCGCCCAUCCACCACCUGCCUAAC----------------------- ...((((.(((((.((.(((((...)))))))......(((....)))))))))))).....................----------------------- ( -13.50, z-score = -0.66, R) >droMoj3.scaffold_6540 18011114 80 - 34148556 UGUCGACAUUUCUAUGACUGAUUUAAUCAGCAGACUUUGACAAGAGCCAGGAAGUCGCCCAACCACCACCUGCCUGACCC--------------------- ((((((........((.(((((...)))))))....))))))..((.((((..((.(.....).))..)))).)).....--------------------- ( -13.00, z-score = -0.30, R) >droGri2.scaffold_14906 3073341 85 - 14172833 UGUCGACAUUUCUAUGACUGAUUUAAUCAGCAGACUUUGACAAGAGCCAGGAAAUCGCCCAUCCACCACCUGACUAACCCCCCUG---------------- ((((((........((.(((((...)))))))....))))))..((.((((.................)))).))..........---------------- ( -11.73, z-score = -0.74, R) >consensus UUUCGACAUUUCUAUGACUGAUUUAAUCAGCAAACUUUGACAAGAGUCAGGAAGUCAACCGUCCACCGUCCGCCAUCCCUC_UCC__C_____________ ....(((.(((((.((.(((((...))))))).(((((....))))).))))))))............................................. (-10.58 = -10.92 + 0.35)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:20:31 2011