
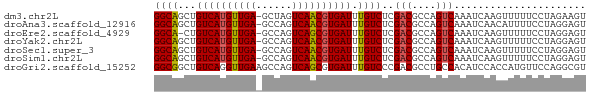
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,664,872 – 7,665,061 |
| Length | 189 |
| Max. P | 0.726364 |

| Location | 7,664,872 – 7,664,943 |
|---|---|
| Length | 71 |
| Sequences | 7 |
| Columns | 72 |
| Reading direction | forward |
| Mean pairwise identity | 91.52 |
| Shannon entropy | 0.17141 |
| G+C content | 0.49472 |
| Mean single sequence MFE | -20.27 |
| Consensus MFE | -16.81 |
| Energy contribution | -16.77 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.594668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7664872 71 + 23011544 GGCAGCUGUCAUGUUGA-GCUAGUCAACGUGAUUUGUCUCGACGCCAGUCAAAUCAAGUUUUUCCUAGAAGU (((((..((((((((((-.....)))))))))))))))..(((....)))...................... ( -18.20, z-score = -1.47, R) >droAna3.scaffold_12916 15613049 71 + 16180835 GGCAGCUGUCAUGUUGA-GCCAGUCAACGUGAUUUGUCUCGACGCCAGUCAAAUCAACAUUUUCCUAGGAGU (((((..((((((((((-.....)))))))))))))))..(((....)))...........(((....))). ( -18.80, z-score = -1.34, R) >droEre2.scaffold_4929 16593840 70 + 26641161 GGCA-CUGUCAUGUUGA-GCCAGUCAGCGUGAUUUGUCUCGACGCCAGUCAAAUCAAGUUUUUCCUAGGAGU ((((-..((((((((((-.....)))))))))).))))..(((....)))...........(((....))). ( -19.20, z-score = -1.50, R) >droYak2.chr2L 17096577 71 - 22324452 GGCAGCUGUCAUGUUGA-GCCAGUCAACGUGAUUUGUCUCGACGCCAGUCAAAUCAAGUUUUUCCUAGGAGU (((((..((((((((((-.....)))))))))))))))..(((....)))...........(((....))). ( -18.80, z-score = -1.38, R) >droSec1.super_3 3179217 71 + 7220098 GGCAGCUGUCAUGUUGA-GCCAGUCAACGUGAUUUGUCUCGACGCCAGUCAAAUCAAGUUUUUCCUAGGAGU (((((..((((((((((-.....)))))))))))))))..(((....)))...........(((....))). ( -18.80, z-score = -1.38, R) >droSim1.chr2L 7462684 71 + 22036055 GGCAGCUGUCAUGUUGA-GCCAGUCAACGUGAUUUGUCUCGACGCCAGUCAAAUCAAGUUUUUCCUAGGAGU (((((..((((((((((-.....)))))))))))))))..(((....)))...........(((....))). ( -18.80, z-score = -1.38, R) >droGri2.scaffold_15252 16762255 72 + 17193109 GGCGGCUGUCAGGUUGAAGCCAGUCAGCGUGAUUUGUCCCGACGCCUGCCACAUCCACCAUGUUCCAGGCGU ((.(((.((((.(((((......))))).))))..))))).(((((((..((((.....))))..))))))) ( -29.30, z-score = -2.50, R) >consensus GGCAGCUGUCAUGUUGA_GCCAGUCAACGUGAUUUGUCUCGACGCCAGUCAAAUCAAGUUUUUCCUAGGAGU ((((...((((((((((......)))))))))).))))..(((....)))...................... (-16.81 = -16.77 + -0.04)


| Location | 7,664,943 – 7,665,061 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 69.87 |
| Shannon entropy | 0.55801 |
| G+C content | 0.40011 |
| Mean single sequence MFE | -25.89 |
| Consensus MFE | -12.22 |
| Energy contribution | -13.10 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.726364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7664943 118 - 23011544 GCUAAUAUUUGACAACGAAUGAUCAAUAGCUAUUUAGAGGAAGAAGAAAAAGUGGACAAAAUAAUUUUCCUCUUUGGUGCACACAUUUCGCUCAGUGUGCCAUCAGCGAAAGGGCACG (((..((((((....)))))).......(((....(((((((((....................)))))))))..(((((((((..........))))).))))))).....)))... ( -26.15, z-score = -0.79, R) >droSim1.chr2L 7462755 115 - 22036055 GCUAAUAUUUGG--AAAAAGUAUCAAUGGCCAUUUACAGGAAAAAGAAAAAGUGGA-AAAAAAAUUUUCCUCUAUGGUGCACACGUUUCGAUCAGUGUGCCACCAGCGAAAGGGCACG ....((((((..--...)))))).....(((...................((.(((-(((....)))))).)).((((((((((..........))))).))))).(....))))... ( -27.40, z-score = -1.44, R) >droSec1.super_3 3179288 117 - 7220098 GCUAAUAUAUGGCAAAAAAGUAUCAAUGGCCAUUUUCAAGAAAACAAAAAAGUGGA-AAAAUAAUUUUCCUCUAUGGUGCACACGUUUCGCUCAGUGUGCCACCAGCGAAAGGGCACG ((((.....))))...............(((.(((((.............((.(((-(((....)))))).)).((((((((((..........))))).)))))..))))))))... ( -32.00, z-score = -2.95, R) >droYak2.chr2L 17096648 95 + 22324452 -----------------------CAAAAACAAUUUAUAGAAAGCAGAAAAAGUGGAAUACAUUUUUUUCCUAAAUGCUGCACACGUUUUGCUCAGUGUGAGACCAACGAAAGGGCAGG -----------------------......................(((((((((.....)))))))))........((((....((((..(.....)..))))...(....).)))). ( -18.80, z-score = -0.69, R) >droEre2.scaffold_4929 16593910 102 - 26641161 UUAGAUAUUUAGCACAAAAGGUUCAAAAGCCACUUA-AGGCAGAUGAAAAAAUG--------UGUUUUCC-------UGUACACGUUUUGCUCAGUGUGACGCCAGCGAAAGGGCACG ....(((((.(((........((((...(((.....-.)))...))))((((((--------(((.....-------...)))))))))))).)))))...(((..(....))))... ( -25.10, z-score = -0.91, R) >consensus GCUAAUAUUUGGCAAAAAAGGAUCAAUAGCCAUUUACAGGAAGAAGAAAAAGUGGA_AAAAUAAUUUUCCUCUAUGGUGCACACGUUUCGCUCAGUGUGCCACCAGCGAAAGGGCACG ............................(((...........................................((((((((((..........))))).))))).(....))))... (-12.22 = -13.10 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:24:10 2011