| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,519,542 – 12,519,634 |
| Length | 92 |
| Max. P | 0.721143 |

| Location | 12,519,542 – 12,519,634 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 76.80 |
| Shannon entropy | 0.44596 |
| G+C content | 0.45525 |
| Mean single sequence MFE | -22.41 |
| Consensus MFE | -15.82 |
| Energy contribution | -15.86 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.14 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.649306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
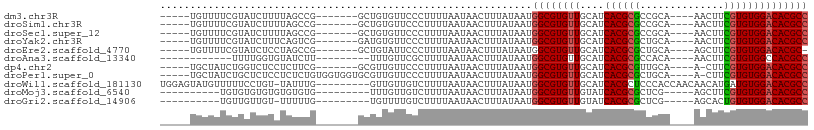
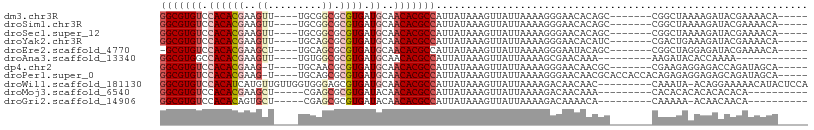
>dm3.chr3R 12519542 92 + 27905053 -----UGUUUUCGUAUCUUUUAGCCG-------GCUGUGUUCCCUUUUAAUAACUUUAUAAUGGCGUGUUGCAUCACGCGCCGCA----AACUUCGUGUGGACACGCC -----....................(-------((.((((((....................(((((((......)))))))(((----.......)))))))))))) ( -22.20, z-score = -0.61, R) >droSim1.chr3R 18566780 92 - 27517382 -----UGUUUUCGUAUCUUUUAGCCG-------GCUGUGUUCCCUUUUAAUAACUUUAUAAUGGCGUGUUGCAUCACGCGCCGCA----AACUUCGUGUGGACACGCC -----....................(-------((.((((((....................(((((((......)))))))(((----.......)))))))))))) ( -22.20, z-score = -0.61, R) >droSec1.super_12 495452 92 - 2123299 -----UGUUUUCGUAUCUUUUAGCCG-------GCUGUGUUCCCUUUUAAUAACUUUAUAAUGGCGUGUUGCAUCACGCGCCGCA----AACUUCGUGUGGACACGCC -----....................(-------((.((((((....................(((((((......)))))))(((----.......)))))))))))) ( -22.20, z-score = -0.61, R) >droYak2.chr3R 16961738 92 - 28832112 -----UGUUUUCGUAUCUUUCAGUCG-------GAUGUGUUCCCUUUUAAUAACUUUAUAAUGGCGUGUUGCAUCACGCGCUGCA----AACUUCGUGUGGACACGCC -----....................(-------((.....)))...................((((((((....((((((..(..----..)..)))))))))))))) ( -20.90, z-score = -0.46, R) >droEre2.scaffold_4770 8634937 91 - 17746568 -----UGUUUUCGUAUCUCCUAGCCG-------GCUGUAUUCCCUUUUAAUAACUUUAUAAUGGCGUGUUGCAUCACGCGCUGCA----AGCUUCGUGUGGACACGC- -----.................((((-------..((((.................)))).))))(((((....((((((..(..----..)..)))))))))))..- ( -18.43, z-score = 0.74, R) >droAna3.scaffold_13340 16332743 83 - 23697760 ------------UUUUGGUGUAUCUU---------UUUGUUCGCUUUUAAUAACUUUAUAAUGGCGUGUUGCAUCACGCGCCACA----AACUUCGUGUGGCCACGCC ------------....(((((.....---------............................(((((......)))))((((((----.......)))))).))))) ( -21.80, z-score = -1.75, R) >dp4.chr2 17502475 91 + 30794189 -----UGCUAUCUGGUCUCCUCUUCG-------GCGUUGUUCCCUUUUAAUAACUUUAUAAUGGCGUGUUGCAUCACGCGUUGCA----A-CUUCGUGUGGACACGCC -----.(((....((...)).....)-------))((((((.......))))))........((((((((....((((((.....----.-...)))))))))))))) ( -22.40, z-score = -1.25, R) >droPer1.super_0 11351196 98 + 11822988 -----UGCUAUCUGCUCUCCUCUCUGUGGUGGUGCGUUGUUCCCUUUUAAUAACUUUAUAAUGGCGUGUUGCAUCACGCGCUGCA----A-CUUCGUGUGGACACGCC -----........((((.((.......)).)).))((((((.......))))))........((((((((....((((((.....----.-...)))))))))))))) ( -24.10, z-score = -0.70, R) >droWil1.scaffold_181130 11561165 98 - 16660200 UGGAGUAUGUUUUUCCUGU-UAUUUG---------GUUGUUGUCUUUUAAUAACUUUAUAAUGGCGUGUUGCAUCACGCUCCCACCAACAACAUGAUGUGGACACGCC .((((.......)))).((-(((..(---------(((((((.....))))))))..)))))(((((((..(((((.(.............).)))))..).)))))) ( -28.62, z-score = -2.62, R) >droMoj3.scaffold_6540 18002989 84 - 34148556 ----------UGUGUGUGUGUGUGUG---------UUUGUUGUCUUUUAAUAACUUUAUAAUGGCGUGUUGUAUCACGCGCUCG-----AGCUUCGUGUGGACACGCC ----------.((((((.((..((.(---------((((((((......)))))........(((((((......))))))).)-----)))..))..)).)))))). ( -23.90, z-score = -1.56, R) >droGri2.scaffold_14906 3065609 83 - 14172833 ----------UGUUGUUGU-UUUUUG---------UGUUUUGUCUUUUAAUAACUUUAUAAUGGCGUGUUGUAUCACGCGCUCG-----AGCACUGUGUGGACACGCC ----------.(((((((.-......---------............)))))))........((((((((....(((((((...-----.))...))))))))))))) ( -19.81, z-score = -0.93, R) >consensus _____UGUUUUCUUAUCUUUUAUCCG_______GCUGUGUUCCCUUUUAAUAACUUUAUAAUGGCGUGUUGCAUCACGCGCUGCA____AACUUCGUGUGGACACGCC ..............................................................((((((((....((((((..............)))))))))))))) (-15.82 = -15.86 + 0.03)
| Location | 12,519,542 – 12,519,634 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 76.80 |
| Shannon entropy | 0.44596 |
| G+C content | 0.45525 |
| Mean single sequence MFE | -21.51 |
| Consensus MFE | -13.54 |
| Energy contribution | -13.88 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.721143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
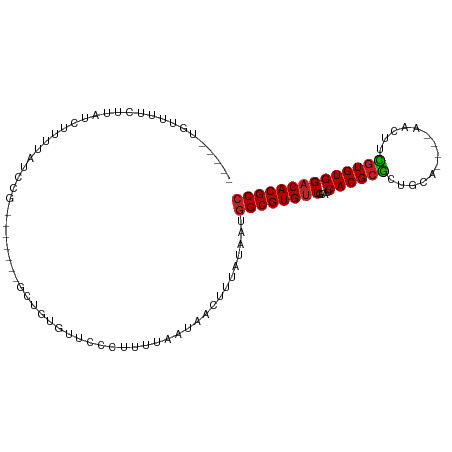
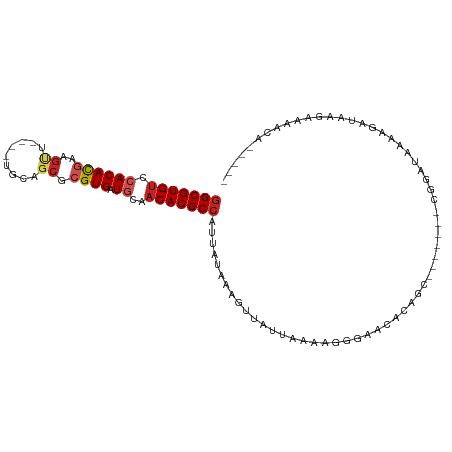
>dm3.chr3R 12519542 92 - 27905053 GGCGUGUCCACACGAAGUU----UGCGGCGCGUGAUGCAACACGCCAUUAUAAAGUUAUUAAAAGGGAACACAGC-------CGGCUAAAAGAUACGAAAACA----- ..((((((.......(((.----.((((((.((......)).))))..................(....)...))-------..)))....))))))......----- ( -20.80, z-score = 0.04, R) >droSim1.chr3R 18566780 92 + 27517382 GGCGUGUCCACACGAAGUU----UGCGGCGCGUGAUGCAACACGCCAUUAUAAAGUUAUUAAAAGGGAACACAGC-------CGGCUAAAAGAUACGAAAACA----- ..((((((.......(((.----.((((((.((......)).))))..................(....)...))-------..)))....))))))......----- ( -20.80, z-score = 0.04, R) >droSec1.super_12 495452 92 + 2123299 GGCGUGUCCACACGAAGUU----UGCGGCGCGUGAUGCAACACGCCAUUAUAAAGUUAUUAAAAGGGAACACAGC-------CGGCUAAAAGAUACGAAAACA----- ..((((((.......(((.----.((((((.((......)).))))..................(....)...))-------..)))....))))))......----- ( -20.80, z-score = 0.04, R) >droYak2.chr3R 16961738 92 + 28832112 GGCGUGUCCACACGAAGUU----UGCAGCGCGUGAUGCAACACGCCAUUAUAAAGUUAUUAAAAGGGAACACAUC-------CGACUGAAAGAUACGAAAACA----- (((((((.((((((..((.----....)).)))).))..)))))))...................(((.....))-------)....................----- ( -19.40, z-score = -0.44, R) >droEre2.scaffold_4770 8634937 91 + 17746568 -GCGUGUCCACACGAAGCU----UGCAGCGCGUGAUGCAACACGCCAUUAUAAAGUUAUUAAAAGGGAAUACAGC-------CGGCUAGGAGAUACGAAAACA----- -.((((((..(....((((----.((.(.(((((......))))))..................(......).))-------.)))).)..))))))......----- ( -20.70, z-score = -0.21, R) >droAna3.scaffold_13340 16332743 83 + 23697760 GGCGUGGCCACACGAAGUU----UGUGGCGCGUGAUGCAACACGCCAUUAUAAAGUUAUUAAAAGCGAACAAA---------AAGAUACACCAAAA------------ ((((((((((((.......----))))))((.....))..))))))........(((......))).......---------..............------------ ( -22.40, z-score = -1.94, R) >dp4.chr2 17502475 91 - 30794189 GGCGUGUCCACACGAAG-U----UGCAACGCGUGAUGCAACACGCCAUUAUAAAGUUAUUAAAAGGGAACAACGC-------CGAAGAGGAGACCAGAUAGCA----- (((((((.((((((..(-(----....)).)))).))..)))))))........((((((....((........)-------)...(.(....)).)))))).----- ( -23.50, z-score = -1.85, R) >droPer1.super_0 11351196 98 - 11822988 GGCGUGUCCACACGAAG-U----UGCAGCGCGUGAUGCAACACGCCAUUAUAAAGUUAUUAAAAGGGAACAACGCACCACCACAGAGAGGAGAGCAGAUAGCA----- (((((((.((((((..(-(----....)).)))).))..)))))))........((((((....(....)...((.(..((.......)).).)).)))))).----- ( -23.90, z-score = -1.40, R) >droWil1.scaffold_181130 11561165 98 + 16660200 GGCGUGUCCACAUCAUGUUGUUGGUGGGAGCGUGAUGCAACACGCCAUUAUAAAGUUAUUAAAAGACAACAAC---------CAAAUA-ACAGGAAAAACAUACUCCA (((((((...(((((((((.(.....).)))))))))..)))))))........((((((.............---------..))))-)).(((.........))). ( -26.86, z-score = -2.40, R) >droMoj3.scaffold_6540 18002989 84 + 34148556 GGCGUGUCCACACGAAGCU-----CGAGCGCGUGAUACAACACGCCAUUAUAAAGUUAUUAAAAGACAACAAA---------CACACACACACACACA---------- (((((((...((((..((.-----...)).)))).....)))))))...........................---------................---------- ( -17.80, z-score = -2.78, R) >droGri2.scaffold_14906 3065609 83 + 14172833 GGCGUGUCCACACAGUGCU-----CGAGCGCGUGAUACAACACGCCAUUAUAAAGUUAUUAAAAGACAAAACA---------CAAAAA-ACAACAACA---------- (((((((...(((.((((.-----...))))))).....)))))))...........................---------......-.........---------- ( -19.60, z-score = -3.10, R) >consensus GGCGUGUCCACACGAAGUU____UGCAGCGCGUGAUGCAACACGCCAUUAUAAAGUUAUUAAAAGGGAACACAGC_______CGGAUAAAAGAUAAGAAAACA_____ (((((((.((((((..(((.......))).)))).))..))))))).............................................................. (-13.54 = -13.88 + 0.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:20:28 2011