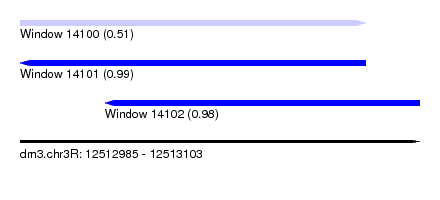
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,512,985 – 12,513,103 |
| Length | 118 |
| Max. P | 0.991929 |

| Location | 12,512,985 – 12,513,087 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.74 |
| Shannon entropy | 0.46424 |
| G+C content | 0.46843 |
| Mean single sequence MFE | -33.69 |
| Consensus MFE | -12.38 |
| Energy contribution | -11.98 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.510635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
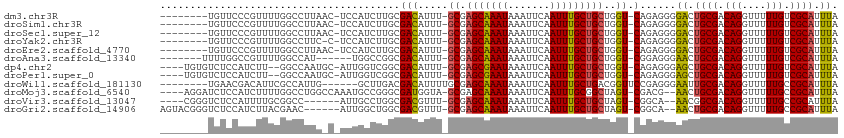
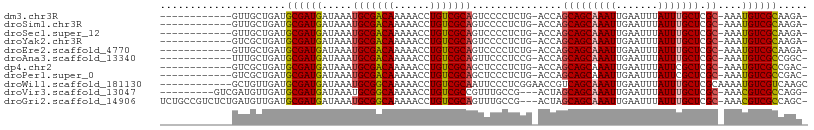
>dm3.chr3R 12512985 102 + 27905053 --------UGUUCCCGUUUUGGCCUUAAC-UCCAUCUUGCGACAUUU-GCGAGCAAAUAAAUUCAAUUUGCUGCUGGU-CAGAGGGGACUGCGACAGGUUUUUGUCGCAUUUA --------.((((((..(((((((.....-.......(((((...))-)))(((((((.......)))))))...)))-))))))))))((((((((....)))))))).... ( -34.70, z-score = -2.97, R) >droSim1.chr3R 18560572 102 - 27517382 --------UGUUCCCGUUUUGGCCUUAAC-UCCAUCUUGCGACAUUU-GCGAGCAAAUAAAUUCAAUUUGCUGCUGGU-CAGAGGGGACUGCGACAGGUUUUUGUCGCAUUUA --------.((((((..(((((((.....-.......(((((...))-)))(((((((.......)))))))...)))-))))))))))((((((((....)))))))).... ( -34.70, z-score = -2.97, R) >droSec1.super_12 488953 102 - 2123299 --------UGUUCCCGUUUUGGCCUUAAC-UCCAUCUUGCGACAUUU-GCGAGCAAAUAAAUUCAAUUUGCUGCUGGU-CAGAGGGGACUGCGACAGGUUUUUGUCGCAUUUA --------.((((((..(((((((.....-.......(((((...))-)))(((((((.......)))))))...)))-))))))))))((((((((....)))))))).... ( -34.70, z-score = -2.97, R) >droYak2.chr3R 16955205 101 - 28832112 --------UGUUCCCGUUUUGGCCUUC-C-UCCAUCUUGCGACAUUU-GCGAGCAAAUAAAUUCAAUUUGCUGCUGGU-CAGAGGGGACUGCGACAGGUUUUUGUCGCAUUUA --------.((((((..(((((((...-.-.......(((((...))-)))(((((((.......)))))))...)))-))))))))))((((((((....)))))))).... ( -34.70, z-score = -2.87, R) >droEre2.scaffold_4770 8628351 102 - 17746568 --------UGUUCCCGUUUUGGCCUUAAC-UCCAUCUUGCGACAUUU-GCGAGCAAAUAAAUUCAAUUUGCUGCUGGU-CAGAGGGGACUGCGACAGGUUUUUGUCGCAUUUA --------.((((((..(((((((.....-.......(((((...))-)))(((((((.......)))))))...)))-))))))))))((((((((....)))))))).... ( -34.70, z-score = -2.97, R) >droAna3.scaffold_13340 16327255 98 - 23697760 -------UUUUGGCCGUUUUGGCCAU------UGGCCGGCGACAUUU-GCGAGCAAAUAAAUUCAAUUUGCUGCUGGU-CGGAGGGAACUGCGACAGGUUUUUGUCGCAUUUA -------...(((((.....))))).------......((((((...-((.(((((((.......)))))))))..((-((.((....)).)))).......))))))..... ( -35.90, z-score = -2.61, R) >dp4.chr2 17495180 104 + 30794189 ----UGUGUCUCCAUCUU--GGCCAAUGC-AUUGGUCGGCGACAUUU-GCGAGCGAAUAAAUUCAAUUUGCUGCUGGU-CAGAGGGAGCUGCGACAGGUUUUUGUCGCAUUUA ----...(.((((.(((.--(((((.(((-(...(((...)))...)-)))(((((((.......)))))))..))))-)))).)))))((((((((....)))))))).... ( -36.20, z-score = -1.87, R) >droPer1.super_0 11343789 104 + 11822988 ----UGUGUCUCCAUCUU--GGCCAAUGC-AUUGGUCGGCGACAUUU-GCGAGCGAAUAAAUUCAAUUUGCUGCUGGU-CAGAGGGAGCUGCGACAGGUUUUUGUCGCAUUUA ----...(.((((.(((.--(((((.(((-(...(((...)))...)-)))(((((((.......)))))))..))))-)))).)))))((((((((....)))))))).... ( -36.20, z-score = -1.87, R) >droWil1.scaffold_181130 11551827 99 - 16660200 --------UGAACGACAUUCGCCAUUG------GCUUGACGACAUUUUGCGAGCAAAUAAAUUCAAUUUGCUGACGGUUCCGAGGGAAUUGCGACAGGUUUUUGCCGCAUUUA --------....((.((...(((...)------)).)).))......((((.(((((.......((((((.((.(((((((...))))))))).))))))))))))))).... ( -22.61, z-score = 0.17, R) >droMoj3.scaffold_6540 17992743 105 - 34148556 ----AGGAUCUCCAUCUUUUGGCCUGGCCAAAUGCCGGGCGAUGGUA-GCGAGCAAAUAAAUUCAAUUUGCGGCUAGU-CGACG--AACUGCGACAGGUUUUUGCCGCAUUUA ----.......(((((.....(((((((.....))))))))))))((-((..((((((.......)))))).))))((-((...--.....)))).(((....)))....... ( -34.70, z-score = -1.22, R) >droVir3.scaffold_13047 13471640 99 - 19223366 ----CGGGUCUCCAUUUUGCGGCC------AUUGCCUGGCGACGUUU-GCGAGCAAAUAAAUUCAAUUUGCUGCUAGU-CGGCA--AACGGCGACAGGUUUUUGCCGCAUUUA ----.((....))....((((((.------(..(((((.((.(((((-((.(((((((.......))))))).(....-.))))--)))).)).)))))..).)))))).... ( -37.50, z-score = -2.50, R) >droGri2.scaffold_14906 3056540 103 - 14172833 AGUACGGGUCUCCAUCUUACGAAC------AUUGGCUGGCGACGUUU-GCGAGCAAAUAAAUUCAAUUUGCUGCUAGU-CGGCA--AACUGCGACAGGUUUUUGCCGCAUUUA .(((.((((....)))))))....------...(((((((..((...-.))(((((((.......)))))))))))))-)((((--((..((.....)).))))))....... ( -27.70, z-score = -0.36, R) >consensus ________UGUCCCCGUUUUGGCCUUA_C_ACUGUCUGGCGACAUUU_GCGAGCAAAUAAAUUCAAUUUGCUGCUGGU_CAGAGGGAACUGCGACAGGUUUUUGUCGCAUUUA ......................................(((.......((.(((((((.......))))))))).........(..((((......))))..)..)))..... (-12.38 = -11.98 + -0.40)
| Location | 12,512,985 – 12,513,087 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.74 |
| Shannon entropy | 0.46424 |
| G+C content | 0.46843 |
| Mean single sequence MFE | -31.37 |
| Consensus MFE | -15.75 |
| Energy contribution | -15.94 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.991929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
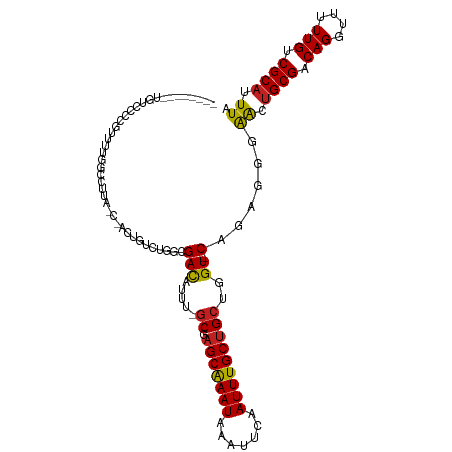
>dm3.chr3R 12512985 102 - 27905053 UAAAUGCGACAAAAACCUGUCGCAGUCCCCUCUG-ACCAGCAGCAAAUUGAAUUUAUUUGCUCGC-AAAUGUCGCAAGAUGGA-GUUAAGGCCAAAACGGGAACA-------- ....(((((((......))))))).((((...((-.((.(((((((((.......))))))).))-....(((....)))...-.....)).))....))))...-------- ( -31.50, z-score = -3.00, R) >droSim1.chr3R 18560572 102 + 27517382 UAAAUGCGACAAAAACCUGUCGCAGUCCCCUCUG-ACCAGCAGCAAAUUGAAUUUAUUUGCUCGC-AAAUGUCGCAAGAUGGA-GUUAAGGCCAAAACGGGAACA-------- ....(((((((......))))))).((((...((-.((.(((((((((.......))))))).))-....(((....)))...-.....)).))....))))...-------- ( -31.50, z-score = -3.00, R) >droSec1.super_12 488953 102 + 2123299 UAAAUGCGACAAAAACCUGUCGCAGUCCCCUCUG-ACCAGCAGCAAAUUGAAUUUAUUUGCUCGC-AAAUGUCGCAAGAUGGA-GUUAAGGCCAAAACGGGAACA-------- ....(((((((......))))))).((((...((-.((.(((((((((.......))))))).))-....(((....)))...-.....)).))....))))...-------- ( -31.50, z-score = -3.00, R) >droYak2.chr3R 16955205 101 + 28832112 UAAAUGCGACAAAAACCUGUCGCAGUCCCCUCUG-ACCAGCAGCAAAUUGAAUUUAUUUGCUCGC-AAAUGUCGCAAGAUGGA-G-GAAGGCCAAAACGGGAACA-------- ....(((((((......)))))))((.(((....-.((.(((((((((.......))))))).))-....(((....))))).-(-(....)).....))).)).-------- ( -32.50, z-score = -3.07, R) >droEre2.scaffold_4770 8628351 102 + 17746568 UAAAUGCGACAAAAACCUGUCGCAGUCCCCUCUG-ACCAGCAGCAAAUUGAAUUUAUUUGCUCGC-AAAUGUCGCAAGAUGGA-GUUAAGGCCAAAACGGGAACA-------- ....(((((((......))))))).((((...((-.((.(((((((((.......))))))).))-....(((....)))...-.....)).))....))))...-------- ( -31.50, z-score = -3.00, R) >droAna3.scaffold_13340 16327255 98 + 23697760 UAAAUGCGACAAAAACCUGUCGCAGUUCCCUCCG-ACCAGCAGCAAAUUGAAUUUAUUUGCUCGC-AAAUGUCGCCGGCCA------AUGGCCAAAACGGCCAAAA------- ....(((((((......)))))))....((..((-((..(((((((((.......))))))).))-....))))..))...------.(((((.....)))))...------- ( -32.80, z-score = -4.14, R) >dp4.chr2 17495180 104 - 30794189 UAAAUGCGACAAAAACCUGUCGCAGCUCCCUCUG-ACCAGCAGCAAAUUGAAUUUAUUCGCUCGC-AAAUGUCGCCGACCAAU-GCAUUGGCC--AAGAUGGAGACACA---- ....(((((((......)))))))(((((.((((-((..(((((.(((.......))).))).))-....)))(((((.....-...))))).--.))).)))).)...---- ( -30.20, z-score = -2.70, R) >droPer1.super_0 11343789 104 - 11822988 UAAAUGCGACAAAAACCUGUCGCAGCUCCCUCUG-ACCAGCAGCAAAUUGAAUUUAUUCGCUCGC-AAAUGUCGCCGACCAAU-GCAUUGGCC--AAGAUGGAGACACA---- ....(((((((......)))))))(((((.((((-((..(((((.(((.......))).))).))-....)))(((((.....-...))))).--.))).)))).)...---- ( -30.20, z-score = -2.70, R) >droWil1.scaffold_181130 11551827 99 + 16660200 UAAAUGCGGCAAAAACCUGUCGCAAUUCCCUCGGAACCGUCAGCAAAUUGAAUUUAUUUGCUCGCAAAAUGUCGUCAAGC------CAAUGGCGAAUGUCGUUCA-------- ....(((((((......))))))).........((((.((.(((((((.......))))))).))......((((((...------...)))))).....)))).-------- ( -27.90, z-score = -2.35, R) >droMoj3.scaffold_6540 17992743 105 + 34148556 UAAAUGCGGCAAAAACCUGUCGCAGUU--CGUCG-ACUAGCCGCAAAUUGAAUUUAUUUGCUCGC-UACCAUCGCCCGGCAUUUGGCCAGGCCAAAAGAUGGAGAUCCU---- ....(((((((......)))))))...--.(((.-..((((.((((((.......))))))..))-))((((((((.(((.....))).))).....))))).)))...---- ( -34.30, z-score = -2.22, R) >droVir3.scaffold_13047 13471640 99 + 19223366 UAAAUGCGGCAAAAACCUGUCGCCGUU--UGCCG-ACUAGCAGCAAAUUGAAUUUAUUUGCUCGC-AAACGUCGCCAGGCAAU------GGCCGCAAAAUGGAGACCCG---- ....((((((.....((((.((.((((--(((..-....(.(((((((.......))))))))))-))))).)).))))....------.))))))....((....)).---- ( -35.30, z-score = -2.93, R) >droGri2.scaffold_14906 3056540 103 + 14172833 UAAAUGCGGCAAAAACCUGUCGCAGUU--UGCCG-ACUAGCAGCAAAUUGAAUUUAUUUGCUCGC-AAACGUCGCCAGCCAAU------GUUCGUAAGAUGGAGACCCGUACU ....(((((((......)))))))(((--(((..-....(.(((((((.......))))))))))-))))(((.(((......------..((....))))).)))....... ( -27.20, z-score = -1.11, R) >consensus UAAAUGCGACAAAAACCUGUCGCAGUUCCCUCUG_ACCAGCAGCAAAUUGAAUUUAUUUGCUCGC_AAAUGUCGCCAGACAGA_G_UAAGGCCAAAACGGGAACA________ ....(((((((......)))))))...............(((((((((.......))))))).))................................................ (-15.75 = -15.94 + 0.19)


| Location | 12,513,010 – 12,513,103 |
|---|---|
| Length | 93 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 87.22 |
| Shannon entropy | 0.24964 |
| G+C content | 0.46669 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -23.12 |
| Energy contribution | -23.05 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
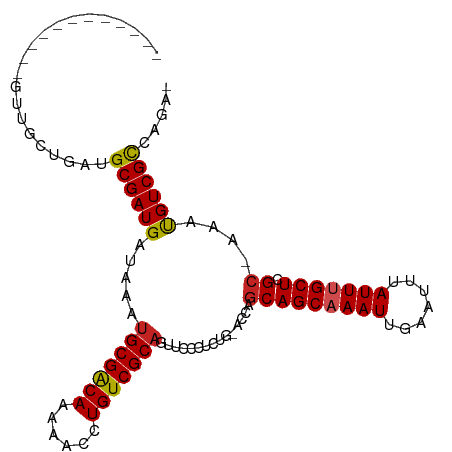
>dm3.chr3R 12513010 93 - 27905053 ------------GUUGCUGAUGCGAUGAUAAAUGCGACAAAAACCUGUCGCAGUCCCCUCUG-ACCAGCAGCAAAUUGAAUUUAUUUGCUCGC-AAAUGUCGCAAGA- ------------........((((((......(((((((......)))))))(((......)-))..(((((((((.......))))))).))-....))))))...- ( -28.00, z-score = -3.07, R) >droSim1.chr3R 18560597 93 + 27517382 ------------GUUGCUGAUGCGAUGAUAAAUGCGACAAAAACCUGUCGCAGUCCCCUCUG-ACCAGCAGCAAAUUGAAUUUAUUUGCUCGC-AAAUGUCGCAAGA- ------------........((((((......(((((((......)))))))(((......)-))..(((((((((.......))))))).))-....))))))...- ( -28.00, z-score = -3.07, R) >droSec1.super_12 488978 93 + 2123299 ------------GUUGCUGAUGCGAUGAUAAAUGCGACAAAAACCUGUCGCAGUCCCCUCUG-ACCAGCAGCAAAUUGAAUUUAUUUGCUCGC-AAAUGUCGCAAGA- ------------........((((((......(((((((......)))))))(((......)-))..(((((((((.......))))))).))-....))))))...- ( -28.00, z-score = -3.07, R) >droYak2.chr3R 16955229 93 + 28832112 ------------GUCGCUGAUGCGAUGAUAAAUGCGACAAAAACCUGUCGCAGUCCCCUCUG-ACCAGCAGCAAAUUGAAUUUAUUUGCUCGC-AAAUGUCGCAAGA- ------------(((((....)))))......(((((((.......((((.((....)).))-))..(((((((((.......))))))).))-...)))))))...- ( -29.00, z-score = -3.31, R) >droEre2.scaffold_4770 8628376 93 + 17746568 ------------GUUGCUGAUGCGAUGAUAAAUGCGACAAAAACCUGUCGCAGUCCCCUCUG-ACCAGCAGCAAAUUGAAUUUAUUUGCUCGC-AAAUGUCGCAAGA- ------------........((((((......(((((((......)))))))(((......)-))..(((((((((.......))))))).))-....))))))...- ( -28.00, z-score = -3.07, R) >droAna3.scaffold_13340 16327276 93 + 23697760 ------------UUUGCUGAUGCGAUGAUAAAUGCGACAAAAACCUGUCGCAGUUCCCUCCG-ACCAGCAGCAAAUUGAAUUUAUUUGCUCGC-AAAUGUCGCCGGC- ------------...((((..(((((......(((((((......)))))))..........-....(((((((((.......))))))).))-....)))))))))- ( -28.80, z-score = -2.87, R) >dp4.chr2 17495207 93 - 30794189 ------------GUCGCUGAUGCGAUGAUAAAUGCGACAAAAACCUGUCGCAGCUCCCUCUG-ACCAGCAGCAAAUUGAAUUUAUUCGCUCGC-AAAUGUCGCCGAC- ------------((((.((((((((((((((((((((((......)))))).(((....((.-...)).))).......))))).))).))))-)....))).))))- ( -24.60, z-score = -1.62, R) >droPer1.super_0 11343816 93 - 11822988 ------------GUCGCUGAUGCGAUGAUAAAUGCGACAAAAACCUGUCGCAGCUCCCUCUG-ACCAGCAGCAAAUUGAAUUUAUUCGCUCGC-AAAUGUCGCCGAC- ------------((((.((((((((((((((((((((((......)))))).(((....((.-...)).))).......))))).))).))))-)....))).))))- ( -24.60, z-score = -1.62, R) >droWil1.scaffold_181130 11551846 96 + 16660200 ------------GCUGUUGAUGCGAUGAUAAAUGCGGCAAAAACCUGUCGCAAUUCCCUCGGAACCGUCAGCAAAUUGAAUUUAUUUGCUCGCAAAAUGUCGUCAAGC ------------((.......))(((((((..(((((((......))))))).((((...))))..((.(((((((.......))))))).))....))))))).... ( -27.90, z-score = -2.24, R) >droVir3.scaffold_13047 13471664 94 + 19223366 ---------GUCGAUGUUGAUGCGAUGAUAAAUGCGGCAAAAACCUGUCGCCGUUUGCCG---ACUAGCAGCAAAUUGAAUUUAUUUGCUCGC-AAACGUCGCCAGG- ---------(.(((((((..(((((.(((((((((((((......)))))).((((((.(---.....).))))))...)))))))...))))-)))))))).)...- ( -29.30, z-score = -1.62, R) >droGri2.scaffold_14906 3056568 103 + 14172833 UCUGCCGUCUCUGAUGUUGAUGCGAUGAUAAAUGCGGCAAAAACCUGUCGCAGUUUGCCG---ACUAGCAGCAAAUUGAAUUUAUUUGCUCGC-AAACGUCGCCAGC- ............((((((..(((((.(((((((((((((......))))))(((((((.(---.....).)))))))..)))))))...))))-)))))))......- ( -29.40, z-score = -1.04, R) >consensus ____________GUUGCUGAUGCGAUGAUAAAUGCGACAAAAACCUGUCGCAGUUCCCUCUG_ACCAGCAGCAAAUUGAAUUUAUUUGCUCGC_AAAUGUCGCCAGA_ .....................((((((.....(((((((......)))))))...............(((((((((.......))))))).))....))))))..... (-23.12 = -23.05 + -0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:20:27 2011