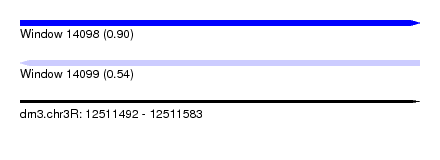
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,511,492 – 12,511,583 |
| Length | 91 |
| Max. P | 0.904626 |

| Location | 12,511,492 – 12,511,583 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 79.37 |
| Shannon entropy | 0.42193 |
| G+C content | 0.33186 |
| Mean single sequence MFE | -15.93 |
| Consensus MFE | -8.06 |
| Energy contribution | -8.22 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.904626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
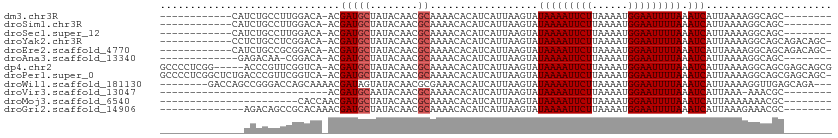
>dm3.chr3R 12511492 91 + 27905053 ------------CAUCUGCCUUGGACA-ACGAUGCUAUACAACGCAAAACACAUCAUUAAGUAUAAAAUUCUUAAAAUGGAAUUUUAAAUCAUUAAAAGGCAGC-------- ------------...(((((((.....-....(((........))).................(((((((((......))))))))).........))))))).-------- ( -18.70, z-score = -3.50, R) >droSim1.chr3R 18559098 91 - 27517382 ------------CAUCUGCCUUGGACA-ACGAUGCUAUACAACGCAAAACACAUCAUUAAGUAUAAAAUUCUUAAAAUGGAAUUUUAAAUCAUUAAAAGGCAGC-------- ------------...(((((((.....-....(((........))).................(((((((((......))))))))).........))))))).-------- ( -18.70, z-score = -3.50, R) >droSec1.super_12 487478 91 - 2123299 ------------CAUCUGCCUUGGACA-ACGAUGCUAUACAACGCAAAACACAUCAUUAAGUAUAAAAUUCUUAAAAUGGAAUUUUAAAUCAUUAAAAGGCAGC-------- ------------...(((((((.....-....(((........))).................(((((((((......))))))))).........))))))).-------- ( -18.70, z-score = -3.50, R) >droYak2.chr3R 16953612 98 - 28832112 ------------CCUCUGCCUCGGACA-ACGAUGCUAUACAACGCAAAACACAUCAUUAAGUAUAAAAUUCUUAAAAUGGAAUUUUAAAUCAUUAAAAGGCAGCAGACAGC- ------------.((((((((((....-.)))(((........))).................(((((((((......)))))))))...........))))).)).....- ( -19.60, z-score = -3.01, R) >droEre2.scaffold_4770 8626825 98 - 17746568 ------------CAUCUGCCGCGGACA-ACGAUGCUAUACAACGCAAAACACAUCAUUAAGUAUAAAAUUCUUAAAAUGGAAUUUUAAAUCAUUAAAAGGCAGCAGACAGC- ------------..(((((.((.....-....(((........))).................(((((((((......)))))))))............)).)))))....- ( -18.80, z-score = -2.57, R) >droAna3.scaffold_13340 16325846 89 - 23697760 -------------GAGACAA-CGGACA-ACGAUGCUAUACAACGCAAAACACAUCAUUAAGUAUAAAAUUCUUAAAAUGGAAUUUUAAAUCAUUAAAAGGCAGC-------- -------------.((.((.-((....-.)).)))).......((.....((........)).(((((((((......)))))))))............))...-------- ( -10.10, z-score = -1.40, R) >dp4.chr2 17493957 106 + 30794189 GCCCCUCGG-----ACCCGUUCGGUCA-ACGAUGCUAUACAACGCAAAACACAUCAUUAAGUAUAAAAUUCUUAAAAUGGAAUUUUAAAUCAUUAAAAGGCAGCGAGCAGCG ((..(((((-----(((.....)))).-....((((..............((........)).(((((((((......)))))))))...........)))).))))..)). ( -22.00, z-score = -1.75, R) >droPer1.super_0 11342479 110 + 11822988 GCCCCUCGGCUCUGACCCGUUCGGUCA-ACGAUGCUAUACAACGCAAAACACAUCAUUAAGUAUAAAAUUCUUAAAAUGGAAUUUUAAAUCAUUAAAAGGCAGCGAGCAGC- ((..(((((((.(((((.....)))))-..(((((........))..................(((((((((......))))))))).))).......)))..))))..))- ( -21.80, z-score = -1.14, R) >droWil1.scaffold_181130 11550408 101 - 16660200 --------GACCAGCCGGGACCAGCAAAACGAUAGUAUACAACGCGAAACACAUCAUUAAGUAUAAAAUUCUUAAAAUGGAAUUUUAAAUCAUUAAAAGGUUGAGCAGA--- --------(..(((((.............((...((.....)).)).................(((((((((......)))))))))...........)))))..)...--- ( -14.80, z-score = -0.91, R) >droVir3.scaffold_13047 13469842 75 - 19223366 ----------------------------ACGAUGCAAUACAACGCAAAACACAUCAUUAAGUAUAAAAUUCUUAAAAUGGAAUUUUAAAUCAUUAAA-AAACGC-------- ----------------------------..(((((........))..................(((((((((......))))))))).)))......-......-------- ( -8.90, z-score = -2.02, R) >droMoj3.scaffold_6540 17991151 81 - 34148556 -----------------------CACCAACGAUGCUAUACAACGCAAAACACAUCAUUAAGUAUAAAAUUCUUAAAAUGGAAUUUUAAAUCAUUAAAAAAACGC-------- -----------------------.......(((((........))..................(((((((((......))))))))).))).............-------- ( -8.80, z-score = -1.98, R) >droGri2.scaffold_14906 3055018 90 - 14172833 --------------AGACAGCCGCACAAACGAUGCUAUACAACGCAAAACACAUCAUUAAGUAUAAAAUUCUUAAAAUGGAAUUUUAAAUCAUUAAAGAAACGC-------- --------------........((........(((........))).......((.((((...(((((((((......))))))))).....)))).))...))-------- ( -10.30, z-score = -1.11, R) >consensus ______________UCUGCCUCGGACA_ACGAUGCUAUACAACGCAAAACACAUCAUUAAGUAUAAAAUUCUUAAAAUGGAAUUUUAAAUCAUUAAAAGGCAGC________ ..............................(((((........))..................(((((((((......))))))))).)))..................... ( -8.06 = -8.22 + 0.17)
| Location | 12,511,492 – 12,511,583 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 79.37 |
| Shannon entropy | 0.42193 |
| G+C content | 0.33186 |
| Mean single sequence MFE | -20.47 |
| Consensus MFE | -9.43 |
| Energy contribution | -9.52 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.541787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
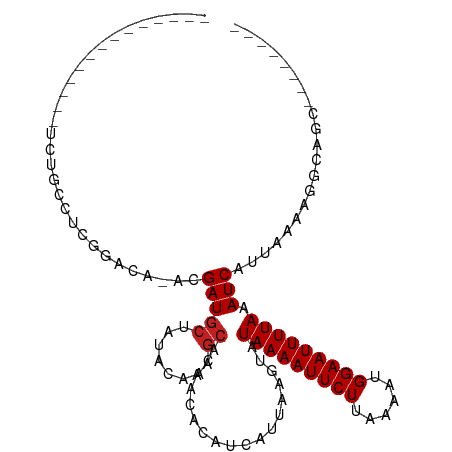
>dm3.chr3R 12511492 91 - 27905053 --------GCUGCCUUUUAAUGAUUUAAAAUUCCAUUUUAAGAAUUUUAUACUUAAUGAUGUGUUUUGCGUUGUAUAGCAUCGU-UGUCCAAGGCAGAUG------------ --------.(((((((.........((((((((........))))))))....(((((((((....(((...)))..)))))))-))...)))))))...------------ ( -22.90, z-score = -3.24, R) >droSim1.chr3R 18559098 91 + 27517382 --------GCUGCCUUUUAAUGAUUUAAAAUUCCAUUUUAAGAAUUUUAUACUUAAUGAUGUGUUUUGCGUUGUAUAGCAUCGU-UGUCCAAGGCAGAUG------------ --------.(((((((.........((((((((........))))))))....(((((((((....(((...)))..)))))))-))...)))))))...------------ ( -22.90, z-score = -3.24, R) >droSec1.super_12 487478 91 + 2123299 --------GCUGCCUUUUAAUGAUUUAAAAUUCCAUUUUAAGAAUUUUAUACUUAAUGAUGUGUUUUGCGUUGUAUAGCAUCGU-UGUCCAAGGCAGAUG------------ --------.(((((((.........((((((((........))))))))....(((((((((....(((...)))..)))))))-))...)))))))...------------ ( -22.90, z-score = -3.24, R) >droYak2.chr3R 16953612 98 + 28832112 -GCUGUCUGCUGCCUUUUAAUGAUUUAAAAUUCCAUUUUAAGAAUUUUAUACUUAAUGAUGUGUUUUGCGUUGUAUAGCAUCGU-UGUCCGAGGCAGAGG------------ -.....((.(((((((.........((((((((........))))))))....(((((((((....(((...)))..)))))))-))...))))))))).------------ ( -24.10, z-score = -2.24, R) >droEre2.scaffold_4770 8626825 98 + 17746568 -GCUGUCUGCUGCCUUUUAAUGAUUUAAAAUUCCAUUUUAAGAAUUUUAUACUUAAUGAUGUGUUUUGCGUUGUAUAGCAUCGU-UGUCCGCGGCAGAUG------------ -...(((((((((............((((((((........))))))))....(((((((((....(((...)))..)))))))-))...))))))))).------------ ( -27.60, z-score = -3.89, R) >droAna3.scaffold_13340 16325846 89 + 23697760 --------GCUGCCUUUUAAUGAUUUAAAAUUCCAUUUUAAGAAUUUUAUACUUAAUGAUGUGUUUUGCGUUGUAUAGCAUCGU-UGUCCG-UUGUCUC------------- --------.........(((((...((((((((........))))))))....(((((((((....(((...)))..)))))))-))..))-)))....------------- ( -14.20, z-score = -1.53, R) >dp4.chr2 17493957 106 - 30794189 CGCUGCUCGCUGCCUUUUAAUGAUUUAAAAUUCCAUUUUAAGAAUUUUAUACUUAAUGAUGUGUUUUGCGUUGUAUAGCAUCGU-UGACCGAACGGGU-----CCGAGGGGC .(((.((((..((((..........((((((((........))))))))...((((((((((....(((...)))..)))))))-)))......))))-----.)))).))) ( -27.40, z-score = -1.46, R) >droPer1.super_0 11342479 110 - 11822988 -GCUGCUCGCUGCCUUUUAAUGAUUUAAAAUUCCAUUUUAAGAAUUUUAUACUUAAUGAUGUGUUUUGCGUUGUAUAGCAUCGU-UGACCGAACGGGUCAGAGCCGAGGGGC -(((.((((((((((..........((((((((........))))))))...((((((((((....(((...)))..)))))))-)))......))).)))...)))).))) ( -28.20, z-score = -1.16, R) >droWil1.scaffold_181130 11550408 101 + 16660200 ---UCUGCUCAACCUUUUAAUGAUUUAAAAUUCCAUUUUAAGAAUUUUAUACUUAAUGAUGUGUUUCGCGUUGUAUACUAUCGUUUUGCUGGUCCCGGCUGGUC-------- ---........(((....((((((.((((((((........))))))))......(..(((((...)))))..).....))))))..((((....)))).))).-------- ( -17.70, z-score = -1.19, R) >droVir3.scaffold_13047 13469842 75 + 19223366 --------GCGUUU-UUUAAUGAUUUAAAAUUCCAUUUUAAGAAUUUUAUACUUAAUGAUGUGUUUUGCGUUGUAUUGCAUCGU---------------------------- --------(((((.-.((((.(...((((((((........))))))))..))))).)))))....((((......))))....---------------------------- ( -9.90, z-score = -0.13, R) >droMoj3.scaffold_6540 17991151 81 + 34148556 --------GCGUUUUUUUAAUGAUUUAAAAUUCCAUUUUAAGAAUUUUAUACUUAAUGAUGUGUUUUGCGUUGUAUAGCAUCGUUGGUG----------------------- --------.((((.....))))...((((((((........))))))))((((.((((((((....(((...)))..))))))))))))----------------------- ( -13.60, z-score = -1.06, R) >droGri2.scaffold_14906 3055018 90 + 14172833 --------GCGUUUCUUUAAUGAUUUAAAAUUCCAUUUUAAGAAUUUUAUACUUAAUGAUGUGUUUUGCGUUGUAUAGCAUCGUUUGUGCGGCUGUCU-------------- --------((...............((((((((........))))))))(((..((((((((....(((...)))..)))))))).)))..)).....-------------- ( -14.30, z-score = -0.16, R) >consensus ________GCUGCCUUUUAAUGAUUUAAAAUUCCAUUUUAAGAAUUUUAUACUUAAUGAUGUGUUUUGCGUUGUAUAGCAUCGU_UGUCCGAGGCAGA______________ .........................((((((((........))))))))......(((((((....(((...)))..)))))))............................ ( -9.43 = -9.52 + 0.09)

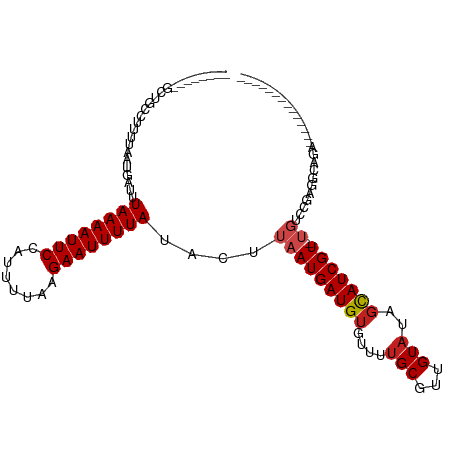
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:20:24 2011