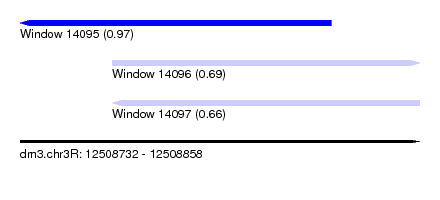
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,508,732 – 12,508,858 |
| Length | 126 |
| Max. P | 0.969832 |

| Location | 12,508,732 – 12,508,830 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 66.39 |
| Shannon entropy | 0.65309 |
| G+C content | 0.47219 |
| Mean single sequence MFE | -26.13 |
| Consensus MFE | -7.89 |
| Energy contribution | -7.75 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.969832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
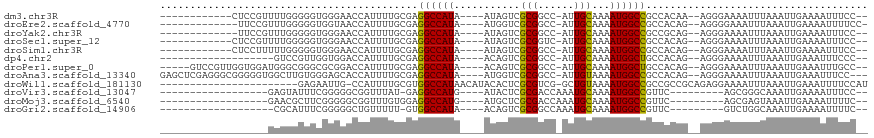
>dm3.chr3R 12508732 98 - 27905053 -CGAGGCCAUA----AUAGUCGCGGCC-AUUGCAAAAUGGCCGCCACAA--AGGGAAAAUUUAAAUU---GAAAAUUUCCGGAAAUACAACCCACCA----CCAUCAUUCCAG -.((((.....----...((.((((((-(((....))))))))).))..--..(((((.(((.....---.))).))))).................----)).))....... ( -25.50, z-score = -2.56, R) >droEre2.scaffold_4770 8624012 83 + 17746568 -CGAGGCCAUA----AUGGUCGCGGCC-AUUGCAAAAUGGCCGCCACAG--AGGGGAAAUUUAAAUU---GAAAAUUUUCCCGAAAACUCGCAC------------------- -((((......----...((.((((((-(((....))))))))).))..--.((((((((((.....---..)))))))))).....))))...------------------- ( -33.60, z-score = -4.57, R) >droYak2.chr3R 16950777 89 + 28832112 -CGAGGCCAUA----AUAGUCGCGGCC-AUUGCAAAAUGGCCGCCGCAG--AGGGAAAAUUUAAAUU---GAAAAUUUCCCGAAAACU--CCCACCAC---CCAG-------- -.(((......----...((.((((((-(((....))))))))).))..--.((((((.(((.....---.))).)))))).....))--).......---....-------- ( -29.80, z-score = -3.99, R) >droSec1.super_12 484688 102 + 2123299 -CGAGGCCAUA----AUAGUCGCGGUC-AUUGCAAAAUGGCCGCCACAG--AGGGAAAAUUUAAAUU---GAAAAUUUCCCGAAAUACAAACCACCACCCACCCCCAUCCCAG -...((.....----...((.((((((-(((....))))))))).))..--.((((((.(((.....---.))).))))))..........)).................... ( -25.50, z-score = -3.08, R) >droSim1.chr3R 18556340 99 + 27517382 -CGAGGCCAUA----AUAGUCGCGGCC-AUUGCAAAAUGGCCGCCACAG--AGGGAAAAUUUAAAUU---GAAAAUUUCCCGAAAUACAAACCACCAC---CACCCACCCCAG -...((.....----...((.((((((-(((....))))))))).))..--.((((((.(((.....---.))).))))))..........)).....---............ ( -28.20, z-score = -4.43, R) >dp4.chr2 17491026 95 - 30794189 -CGAGGCCAUA----ACAGUCGCGGCC-AUUGCAAAAUGGCUGCCACAG--AGGGAAAAUUUAAAUU---GAAAUUUCCCAGAACUCCCCAGAACUCCUGCCACCC------- -...(((....----...((.((((((-(((....))))))))).))..--.((((((.(((.....---))).))))))...................)))....------- ( -29.40, z-score = -3.15, R) >droPer1.super_0 11339326 85 - 11822988 -CGAGGCCAUA----ACAGUCGCGGCC-AUUGCAAAAUGGCUGCCACAG--AGGGAAAAUUUAAAUU---GAAAUUUGCCAGCACUCCUGCCACCC----------------- -...(((....----...((.((((((-(((....))))))))).)).(--..((.((((((.....---.)))))).))..)......)))....----------------- ( -26.00, z-score = -1.76, R) >droAna3.scaffold_13340 16323201 82 + 23697760 -CGAGGCCAUA----AUGGUCGCGGCC-AUUGUAAAAUGGCCGCCACAG--AGGGAAAAUUUAAAUU---GAAAUUUCCCAAAACUCUCGCCG-------------------- -...(((....----...((.((((((-(((....))))))))).))..--.((((((.(((.....---))).)))))).........))).-------------------- ( -31.70, z-score = -4.42, R) >droWil1.scaffold_181130 11546983 95 + 16660200 -CGUGGCCAUAACAUACACUCGCGUCG-GCUGUAAAAUGGCCGCCGCCGCAGAGGAAAAUUUAAAUU---GAAAUUUUCCAUCCAAAAAUUUCCCCUAAA------------- -.((((((((....((((..((...))-..))))..)))))))).........(((((((((.....---.)))))))))....................------------- ( -24.40, z-score = -2.15, R) >droVir3.scaffold_13047 13466033 89 + 19223366 -UGAGGCCAUG----AUACUCGCGACCAAAUGCAAAAUGGCCGUU-CAGC-GGGCAAAUUGAAAAUUUCCCUAAAUUUCCACAGCCGUUCUUGAUG----------------- -...(((((((----.....)(((......)))...))))))(..-(.((-.((.(((((.............)))))))...)).)..)......----------------- ( -15.22, z-score = 1.21, R) >droMoj3.scaffold_6540 17987454 93 + 34148556 UGGAGGCCAUG----AUGCUCGCGACCAAAUGCAAAAUGGCCGUU-CAGC-GAGUAAAUUGAAAAUUUUCCAAAAUUCUCGCAGCCAUUGCUCAGCUCG-------------- (((.((((((.----.(((............)))..))))))...-..((-(((....(((.........)))....)))))..)))............-------------- ( -24.40, z-score = -0.69, R) >droGri2.scaffold_14906 3051714 92 + 14172833 -UGUGGCCAUA----ACAGUCGCGGCCAAAUGCAAAAUGGCCGUU-CGUC-UGGCAAAUUGAAAAUUUUCCAAAAUUUCCACAACUACCGUAUAUCUAU-------------- -(((((.....----.(((.((((((((.........))))))..-)).)-)).........((((((....)))))))))))................-------------- ( -19.90, z-score = -1.18, R) >consensus _CGAGGCCAUA____AUAGUCGCGGCC_AUUGCAAAAUGGCCGCCACAG__AGGGAAAAUUUAAAUU___GAAAAUUUCCAGAAAUACAGCCCACC_________________ ....((((((..........................))))))....................................................................... ( -7.89 = -7.75 + -0.14)
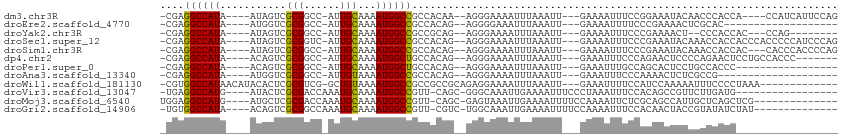
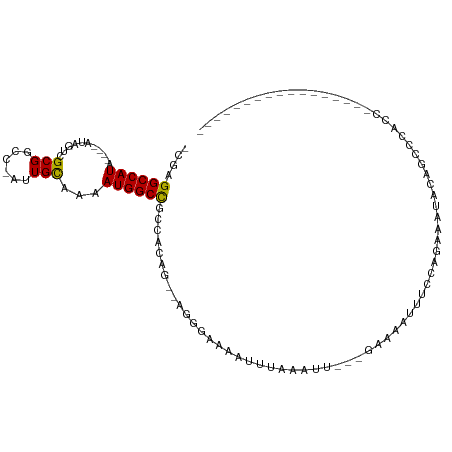

| Location | 12,508,761 – 12,508,858 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 67.86 |
| Shannon entropy | 0.62415 |
| G+C content | 0.48392 |
| Mean single sequence MFE | -27.94 |
| Consensus MFE | -8.45 |
| Energy contribution | -8.78 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.688987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12508761 97 + 27905053 --GGAAAUUUUCAAUUUAAAUUUUCCCU--UUGUGGCGGCCAUUUUGCAAU-GGCCGCGACUAU----UAUGGCCUCGCAAAAUGGUUCCCACCCCCAAAACGGAG------------ --(((((.(((......))).)))))..--..((((..((((((((((...-(((((..(...)----..)))))..))))))))))..))))..((.....))..------------ ( -33.50, z-score = -3.36, R) >droEre2.scaffold_4770 8624025 97 - 17746568 -GGAAAAUUUUCAAUUUAAAUUUCCCCU--CUGUGGCGGCCAUUUUGCAAU-GGCCGCGACCAU----UAUGGCCUCGCAAAAUGGUUACCACCCCCAAACGGAA------------- -(((((.((........)).)))))...--..((((..((((((((((...-(((((..(...)----..)))))..))))))))))..))))..((....))..------------- ( -33.70, z-score = -3.30, R) >droYak2.chr3R 16950797 96 - 28832112 --GGAAAUUUUCAAUUUAAAUUUUCCCU--CUGCGGCGGCCAUUUUGCAAU-GGCCGCGACUAU----UAUGGCCUCGCAAAAUGGUUCCCACCCCCAAACGGAA------------- --(((((.(((......))).)))))..--....((..((((((((((...-(((((..(...)----..)))))..))))))))))..))....((....))..------------- ( -30.40, z-score = -2.24, R) >droSec1.super_12 484721 97 - 2123299 --GGAAAUUUUCAAUUUAAAUUUUCCCU--CUGUGGCGGCCAUUUUGCAAU-GACCGCGACUAU----UAUGGCCUCGCAAAAUGGUUCCCACCCCCAAAACGGAG------------ --(((((.(((......))).)))))((--((((((..((((((((((...-(.(((..(...)----..))).)..))))))))))..)))).........))))------------ ( -26.60, z-score = -2.10, R) >droSim1.chr3R 18556370 97 - 27517382 --GGAAAUUUUCAAUUUAAAUUUUCCCU--CUGUGGCGGCCAUUUUGCAAU-GGCCGCGACUAU----UAUGGCCUCGCAAAAUGGUUCCCACCCCCAAAAAGGAG------------ --(((((.(((......))).)))))((--((((((..((((((((((...-(((((..(...)----..)))))..))))))))))..)))).........))))------------ ( -33.70, z-score = -3.45, R) >dp4.chr2 17491052 90 + 30794189 --GGGAAAUUUCAAUUUAAAUUUUCCCU--CUGUGGCAGCCAUUUUGCAAU-GGCCGCGACUGU----UAUGGCCUCGCAAAAUGGUCCACCAACGGAC------------------- --((((((.............))))))(--((((((..((((((((((...-(((((..(...)----..)))))..))))))))))...)).))))).------------------- ( -34.22, z-score = -3.90, R) >droPer1.super_0 11339342 104 + 11822988 --GGCAAAUUUCAAUUUAAAUUUUCCCU--CUGUGGCAGCCAUUUUGCAAU-GGCCGCGACUGU----UAUGGCCUCGCAAAAUGGUCCGCGCCCGCCCAUCCACCAACGGAC----- --(((((((((......)))))).....--..((((..((((((((((...-(((((..(...)----..)))))..)))))))))))))))))......(((......))).----- ( -32.90, z-score = -1.94, R) >droAna3.scaffold_13340 16323215 108 - 23697760 ---GGAAAUUUCAAUUUAAAUUUUCCCU--CUGUGGCGGCCAUUUUACAAU-GGCCGCGACCAU----UAUGGCCUCGCAAAAUGGUGCUCCCACAAGCCACCCCCGCCCUCGAGCUC ---((((((((......))))))))...--..((((((((((((....)))-))))((.(((((----(.((......)).))))))))........)))))....((......)).. ( -32.30, z-score = -2.36, R) >droWil1.scaffold_181130 11547001 93 - 16660200 AUGGAAAAUUUCAAUUUAAAUUUUCCUCUGCGGCGGCGGCCAUUUUACAGC-CGACGCGAGUGUAUGUUAUGGCCACGCAAAAUGG-CAAUUCUC----------------------- ..(((((((((......)))))))))..(((.(((..((((((...(((..-(.((....)))..))).)))))).)))......)-))......----------------------- ( -28.30, z-score = -1.87, R) >droVir3.scaffold_13047 13466059 84 - 19223366 --GGAAAUUUUCAAUUUGCCCGCU---------GAACGGCCAUUUUGCAUUUGGUCGCGAGUAU----CAUGGCCUC-AUAAACCGCCCCCGAAAUACUC------------------ --.........(((..((.(((..---------...))).))..)))..(((((..(((.((..----.(((....)-))..)))))..)))))......------------------ ( -15.60, z-score = 0.72, R) >droMoj3.scaffold_6540 17987483 85 - 34148556 --GAAAAUUUUCAAUUUACUCGCU---------GAACGGCCAUUUUGCAUUUGGUCGCGAGCAU----CAUGGCCUCCACAAACCGCCCCCGAAGCGUUC------------------ --..................((((---------....((((((..(((..(((....)))))).----.)))))).........((....)).))))...------------------ ( -17.10, z-score = 0.25, R) >droGri2.scaffold_14906 3051743 83 - 14172833 --GAAAAUUUUCAAUUUGCCAGAC---------GAACGGCCAUUUUGCAUUUGGCCGCGACUGU----UAUGGCCAC-AAAAACAGCCCCCGAAAUGCG------------------- --......((((.......(((.(---------(..((((((.........)))))))).))).----...(((...-.......)))...))))....------------------- ( -17.00, z-score = 0.01, R) >consensus __GGAAAUUUUCAAUUUAAAUUUUCCCU__CUGUGGCGGCCAUUUUGCAAU_GGCCGCGACUAU____UAUGGCCUCGCAAAAUGGUCCCCACCCCCAAA__________________ .....................................((((((.((((........)))).........))))))........................................... ( -8.45 = -8.78 + 0.33)

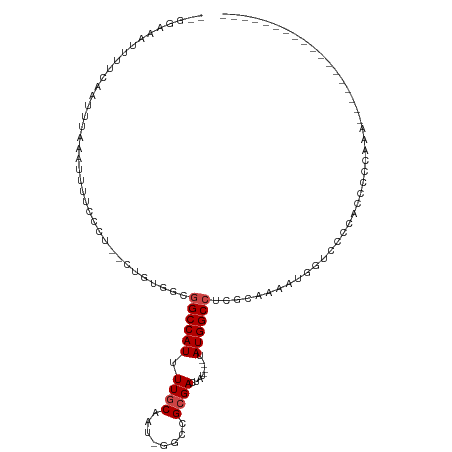

| Location | 12,508,761 – 12,508,858 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 67.86 |
| Shannon entropy | 0.62415 |
| G+C content | 0.48392 |
| Mean single sequence MFE | -33.39 |
| Consensus MFE | -7.89 |
| Energy contribution | -7.75 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.24 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.663728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12508761 97 - 27905053 ------------CUCCGUUUUGGGGGUGGGAACCAUUUUGCGAGGCCAUA----AUAGUCGCGGCC-AUUGCAAAAUGGCCGCCACAA--AGGGAAAAUUUAAAUUGAAAAUUUCC-- ------------.(((.(((((..(((((...(((((((((((((((...----........))))-.)))))))))))))))).)))--))))).....................-- ( -36.40, z-score = -3.08, R) >droEre2.scaffold_4770 8624025 97 + 17746568 -------------UUCCGUUUGGGGGUGGUAACCAUUUUGCGAGGCCAUA----AUGGUCGCGGCC-AUUGCAAAAUGGCCGCCACAG--AGGGGAAAUUUAAAUUGAAAAUUUUCC- -------------((((.((((..(((((...(((((((((((((((...----........))))-.)))))))))))))))).)))--).)))).....................- ( -38.90, z-score = -3.41, R) >droYak2.chr3R 16950797 96 + 28832112 -------------UUCCGUUUGGGGGUGGGAACCAUUUUGCGAGGCCAUA----AUAGUCGCGGCC-AUUGCAAAAUGGCCGCCGCAG--AGGGAAAAUUUAAAUUGAAAAUUUCC-- -------------((((.((((.((.(((...(((((((((((((((...----........))))-.)))))))))))))))).)))--).))))....................-- ( -37.70, z-score = -3.15, R) >droSec1.super_12 484721 97 + 2123299 ------------CUCCGUUUUGGGGGUGGGAACCAUUUUGCGAGGCCAUA----AUAGUCGCGGUC-AUUGCAAAAUGGCCGCCACAG--AGGGAAAAUUUAAAUUGAAAAUUUCC-- ------------.(((.(((((..(((((...(((((((((((((((...----........))))-.)))))))))))))))).)))--))))).....................-- ( -33.80, z-score = -2.71, R) >droSim1.chr3R 18556370 97 + 27517382 ------------CUCCUUUUUGGGGGUGGGAACCAUUUUGCGAGGCCAUA----AUAGUCGCGGCC-AUUGCAAAAUGGCCGCCACAG--AGGGAAAAUUUAAAUUGAAAAUUUCC-- ------------.(((((((.(..(((((...(((((((((((((((...----........))))-.)))))))))))))))).)))--))))).....................-- ( -36.50, z-score = -3.10, R) >dp4.chr2 17491052 90 - 30794189 -------------------GUCCGUUGGUGGACCAUUUUGCGAGGCCAUA----ACAGUCGCGGCC-AUUGCAAAAUGGCUGCCACAG--AGGGAAAAUUUAAAUUGAAAUUUCCC-- -------------------.((.((.((..(.(((((((((((((((...----........))))-.))))))))))))..)))).)--)((((((.(((.....))).))))))-- ( -37.30, z-score = -4.42, R) >droPer1.super_0 11339342 104 - 11822988 -----GUCCGUUGGUGGAUGGGCGGGCGCGGACCAUUUUGCGAGGCCAUA----ACAGUCGCGGCC-AUUGCAAAAUGGCUGCCACAG--AGGGAAAAUUUAAAUUGAAAUUUGCC-- -----(((((((....))))))).(((((((.(((((((((((((((...----........))))-.))))))))))))))).....--.....((((((......)))))))))-- ( -40.20, z-score = -2.26, R) >droAna3.scaffold_13340 16323215 108 + 23697760 GAGCUCGAGGGCGGGGGUGGCUUGUGGGAGCACCAUUUUGCGAGGCCAUA----AUGGUCGCGGCC-AUUGUAAAAUGGCCGCCACAG--AGGGAAAAUUUAAAUUGAAAUUUCC--- ...(((...(((.(..((((((((..(((......)))..).))))))).----.).)))((((((-(((....)))))))))....)--))(((((.(((.....))).)))))--- ( -41.80, z-score = -2.65, R) >droWil1.scaffold_181130 11547001 93 + 16660200 -----------------------GAGAAUUG-CCAUUUUGCGUGGCCAUAACAUACACUCGCGUCG-GCUGUAAAAUGGCCGCCGCCGCAGAGGAAAAUUUAAAUUGAAAUUUUCCAU -----------------------........-....((((((((...........)))..(((.((-(((((...))))))).))).)))))(((((((((......))))))))).. ( -27.90, z-score = -2.00, R) >droVir3.scaffold_13047 13466059 84 + 19223366 ------------------GAGUAUUUCGGGGGCGGUUUAU-GAGGCCAUG----AUACUCGCGACCAAAUGCAAAAUGGCCGUUC---------AGCGGGCAAAUUGAAAAUUUCC-- ------------------..((((((.((..(((((((((-(....))))----).)).)))..))))))))....((.(((...---------..))).))..............-- ( -23.90, z-score = -0.75, R) >droMoj3.scaffold_6540 17987483 85 + 34148556 ------------------GAACGCUUCGGGGGCGGUUUGUGGAGGCCAUG----AUGCUCGCGACCAAAUGCAAAAUGGCCGUUC---------AGCGAGUAAAUUGAAAAUUUUC-- ------------------...((((.....)))).((((((((((((((.----.(((............)))..)))))).)))---------).))))................-- ( -22.20, z-score = 0.41, R) >droGri2.scaffold_14906 3051743 83 + 14172833 -------------------CGCAUUUCGGGGGCUGUUUUU-GUGGCCAUA----ACAGUCGCGGCCAAAUGCAAAAUGGCCGUUC---------GUCUGGCAAAUUGAAAAUUUUC-- -------------------....((((((.(((..(....-)..)))...----.(((.((((((((.........))))))..)---------).))).....))))))......-- ( -24.10, z-score = -0.78, R) >consensus __________________UUUGGGGGUGGGGACCAUUUUGCGAGGCCAUA____AUAGUCGCGGCC_AUUGCAAAAUGGCCGCCACAG__AGGGAAAAUUUAAAUUGAAAAUUUCC__ ...........................................((((((..........................))))))..................................... ( -7.89 = -7.75 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:20:23 2011