
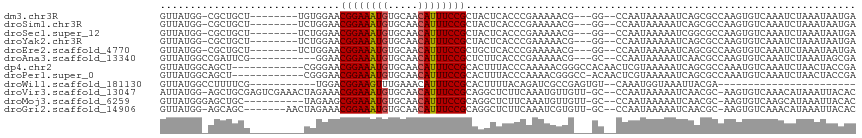
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,490,787 – 12,490,889 |
| Length | 102 |
| Max. P | 0.995553 |

| Location | 12,490,787 – 12,490,889 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 71.67 |
| Shannon entropy | 0.59015 |
| G+C content | 0.43749 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -9.93 |
| Energy contribution | -9.94 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12490787 102 + 27905053 GUUAUGG-CGCUGCU--------UGUGGAACGGAAAUGUGCAACAUUUCCGCUACUCACCCGAAAAACG---GG--CCAAUAAAAAUCAGCGCCAAGUGUCAAAUCUAAAUAAUGA ....(((-(((((.(--------(.(((..((((((((.....)))))))).......((((.....))---))--))).....)).))))))))..................... ( -33.80, z-score = -3.94, R) >droSim1.chr3R 18536794 102 - 27517382 GUUAUGG-CGCUGCU--------UCUGGAACGGAAAUGUGCAACAUUUCCGCUACUCACCCGAAAAACG---GG--CCAAUAAAAAUCAGCGCCAAGUGUCAAAUCUAAAUAAUGA ....(((-(((((..--------..(((..((((((((.....)))))))).......((((.....))---))--)))........))))))))..................... ( -31.80, z-score = -3.67, R) >droSec1.super_12 466757 102 - 2123299 GUUAUGG-CGCUGCU--------UCUGGAACGGAAAUGUGCAACAUUUCCGCUACUCACCCGAAAAACG---GG--CCAAUAAAAAUCGGCGCCAAGUGUCAAAUCUAAAUAAUGA ....(((-(((((..--------..(((..((((((((.....)))))))).......((((.....))---))--)))........))))))))..................... ( -31.10, z-score = -3.07, R) >droYak2.chr3R 16932321 102 - 28832112 GUUAUGG-CGCUGCU--------UCUGGAACGGAAAUGUGCAACAUUUCCGCUACUCACCCGAAAAACG---GG--CCAAUAAAAAUCAGCGCCAAGUGUCAAAUCUAAAUAAUGA ....(((-(((((..--------..(((..((((((((.....)))))))).......((((.....))---))--)))........))))))))..................... ( -31.80, z-score = -3.67, R) >droEre2.scaffold_4770 8605950 102 - 17746568 GUUAUGG-CGCUGCU--------UCUGGAACGGAAAUGUGCAACAUUUCCGCUGCUCACCCGAAAAACG---GG--CCAAUAAAAAUCAGCGCCAAGUGUCAAAUCUAAAUAAUGA ....(((-(((((..--------..(((..((((((((.....)))))))).......((((.....))---))--)))........))))))))..................... ( -31.80, z-score = -3.13, R) >droAna3.scaffold_13340 16306227 100 - 23697760 GUUAUGGCCGAUUCG-----------GGAACGGAAAUGUGCAACAUUUCCGCUCUUCACCCGAAAAACG---GC--CCAAUAAAAAUCAACGCCAAGUGUCAAAUCUAAAUAGCGA ((((((((((.((((-----------((..((((((((.....)))))))).......))))))...))---))--)............((((...)))).........))))).. ( -30.00, z-score = -3.24, R) >dp4.chr2 17474257 104 + 30794189 GUUAUGGCAGCU------------CGGGAACGGAAAUGUGCAACAUUUCCGCACUUUACCCAAAAACGGGCCACAACUCGUAAAAAUCAGCGCCAAAUGUCAAAUCUAACUACCGA ....((((.(((------------.(((((((((((((.....))))))))....)).)))....(((((......))))).......)))))))..................... ( -26.60, z-score = -2.07, R) >droPer1.super_0 11322128 103 + 11822988 GUUAUGGCAGCU------------CGGGAACGGAAAUGUGCAACAUUUCCGCACUUUACCCAAAACGGGCC-ACAACUCGUAAAAAUCAGCGCCAAAUGUCAAAUCUAACUACCGA ....((((.(((------------.(((((((((((((.....))))))))....)).)))...(((((..-....))))).......)))))))..................... ( -26.60, z-score = -2.04, R) >droWil1.scaffold_181130 11525689 80 - 16660200 GUUAUGGCCUUUUCG-----------UGGACGGAAGUUUGAAACAUUUCCGCACUUUUACAGAUCGCCGAGUGU--CAAAUGGUAAAUUACGA----------------------- ....((((..(((.(-----------(((((((((((.......)))))))....))))))))..)))).....--.................----------------------- ( -13.90, z-score = 1.09, R) >droVir3.scaffold_13047 13446516 111 - 19223366 AUUAUGG-AGCUGCGAGUCGAAACUAGAAACGGAAAUGUGCAACAUUUCCGCAGGCUCUUCAAAUGUUGUU-GC--CCAAUAAAAAUCAACGC-AAGUGUCAAACAUAAAUUACAC .((((((-(((((((..((.......))..)(((((((.....)))))))))).))))......(((((..-..--.))))).......((..-..))......)))))....... ( -25.60, z-score = -1.51, R) >droMoj3.scaffold_6259 5991 102 + 16918 GUUAUGGGAGCUGC----------UAGAAGCGGAAAUGUGCAACAUUUCCGCAGGCUCUUCAAAUGUUGUU-GC--CCAAUAAAAAUCAACGC-AAGUGUCAAGCAUAAAUUACAC .(((((((((((..----------.....(((((((((.....))))))))).)))))).....(((((..-..--.))))).......((..-..))......)))))....... ( -26.10, z-score = -1.26, R) >droGri2.scaffold_14906 3031967 104 - 14172833 GUUAUGG-AGCAGC-------AACUAGAAACGGAAAUGUGCAACAUUUCCGCAGGCUCUUCAAAUCGUGUU-GC--CCAAUAAAAAUCAACGC-AAGUGUCAAACAUAAAUUACAC (((.(((-.(((((-------(....((..((((((((.....))))))))..((....))...)).))))-))--)))..........((..-..))....)))........... ( -24.10, z-score = -2.05, R) >consensus GUUAUGG_CGCUGCU_________CUGGAACGGAAAUGUGCAACAUUUCCGCAACUCACCCGAAAAACG___GC__CCAAUAAAAAUCAGCGCCAAGUGUCAAAUCUAAAUAACGA ..............................((((((((.....))))))))................................................................. ( -9.93 = -9.94 + 0.01)


| Location | 12,490,787 – 12,490,889 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 71.67 |
| Shannon entropy | 0.59015 |
| G+C content | 0.43749 |
| Mean single sequence MFE | -31.94 |
| Consensus MFE | -12.10 |
| Energy contribution | -11.57 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.995553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
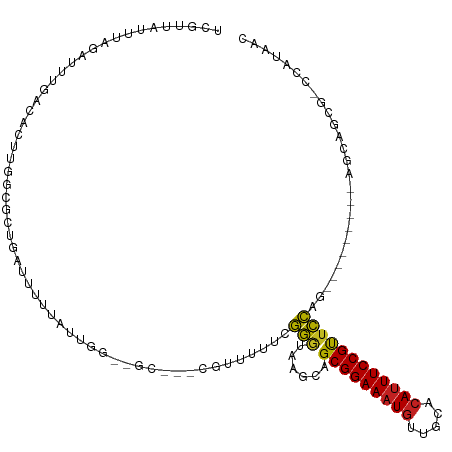
>dm3.chr3R 12490787 102 - 27905053 UCAUUAUUUAGAUUUGACACUUGGCGCUGAUUUUUAUUGG--CC---CGUUUUUCGGGUGAGUAGCGGAAAUGUUGCACAUUUCCGUUCCACA--------AGCAGCG-CCAUAAC .....................((((((((.....((((.(--((---((.....))))).))))(((((((((.....)))))))))......--------..)))))-))).... ( -39.20, z-score = -5.37, R) >droSim1.chr3R 18536794 102 + 27517382 UCAUUAUUUAGAUUUGACACUUGGCGCUGAUUUUUAUUGG--CC---CGUUUUUCGGGUGAGUAGCGGAAAUGUUGCACAUUUCCGUUCCAGA--------AGCAGCG-CCAUAAC .....................((((((((.((((((((.(--((---((.....))))).)))((((((((((.....))))))))))..)))--------)))))))-))).... ( -39.60, z-score = -5.34, R) >droSec1.super_12 466757 102 + 2123299 UCAUUAUUUAGAUUUGACACUUGGCGCCGAUUUUUAUUGG--CC---CGUUUUUCGGGUGAGUAGCGGAAAUGUUGCACAUUUCCGUUCCAGA--------AGCAGCG-CCAUAAC .....................((((((.(.((((((((.(--((---((.....))))).)))((((((((((.....))))))))))..)))--------))).)))-))).... ( -35.00, z-score = -3.86, R) >droYak2.chr3R 16932321 102 + 28832112 UCAUUAUUUAGAUUUGACACUUGGCGCUGAUUUUUAUUGG--CC---CGUUUUUCGGGUGAGUAGCGGAAAUGUUGCACAUUUCCGUUCCAGA--------AGCAGCG-CCAUAAC .....................((((((((.((((((((.(--((---((.....))))).)))((((((((((.....))))))))))..)))--------)))))))-))).... ( -39.60, z-score = -5.34, R) >droEre2.scaffold_4770 8605950 102 + 17746568 UCAUUAUUUAGAUUUGACACUUGGCGCUGAUUUUUAUUGG--CC---CGUUUUUCGGGUGAGCAGCGGAAAUGUUGCACAUUUCCGUUCCAGA--------AGCAGCG-CCAUAAC .....................((((((((.(((((..(.(--((---((.....))))).)(.((((((((((.....)))))))))).))))--------)))))))-))).... ( -38.70, z-score = -4.82, R) >droAna3.scaffold_13340 16306227 100 + 23697760 UCGCUAUUUAGAUUUGACACUUGGCGUUGAUUUUUAUUGG--GC---CGUUUUUCGGGUGAAGAGCGGAAAUGUUGCACAUUUCCGUUCC-----------CGAAUCGGCCAUAAC .(((((...............))))).............(--((---((...((((((.....((((((((((.....))))))))))))-----------)))).)))))..... ( -36.16, z-score = -3.66, R) >dp4.chr2 17474257 104 - 30794189 UCGGUAGUUAGAUUUGACAUUUGGCGCUGAUUUUUACGAGUUGUGGCCCGUUUUUGGGUAAAGUGCGGAAAUGUUGCACAUUUCCGUUCCCG------------AGCUGCCAUAAC (((((.(((((((.....)))))))))))).........((((((((..((..(((((......(((((((((.....))))))))).))))------------))).)))))))) ( -35.80, z-score = -3.09, R) >droPer1.super_0 11322128 103 - 11822988 UCGGUAGUUAGAUUUGACAUUUGGCGCUGAUUUUUACGAGUUGU-GGCCCGUUUUGGGUAAAGUGCGGAAAUGUUGCACAUUUCCGUUCCCG------------AGCUGCCAUAAC (((((.(((((((.....)))))))))))).........(((((-(((..((((.(((......(((((((((.....))))))))).))))------------))).)))))))) ( -36.40, z-score = -3.24, R) >droWil1.scaffold_181130 11525689 80 + 16660200 -----------------------UCGUAAUUUACCAUUUG--ACACUCGGCGAUCUGUAAAAGUGCGGAAAUGUUUCAAACUUCCGUCCA-----------CGAAAAGGCCAUAAC -----------------------((((..(((((...(((--........)))...))))).(..(((((...........)))))..))-----------)))............ ( -12.20, z-score = 0.65, R) >droVir3.scaffold_13047 13446516 111 + 19223366 GUGUAAUUUAUGUUUGACACUU-GCGUUGAUUUUUAUUGG--GC-AACAACAUUUGAAGAGCCUGCGGAAAUGUUGCACAUUUCCGUUUCUAGUUUCGACUCGCAGCU-CCAUAAU .......(((((........((-((.(..((....))..)--))-))...........((((.((((((((((.....)))))))(..((.......))..)))))))-)))))). ( -24.20, z-score = -0.18, R) >droMoj3.scaffold_6259 5991 102 - 16918 GUGUAAUUUAUGCUUGACACUU-GCGUUGAUUUUUAUUGG--GC-AACAACAUUUGAAGAGCCUGCGGAAAUGUUGCACAUUUCCGCUUCUA----------GCAGCUCCCAUAAC ((((............))))((-((...((........((--((-...............))))(((((((((.....))))))))).))..----------)))).......... ( -23.16, z-score = -0.39, R) >droGri2.scaffold_14906 3031967 104 + 14172833 GUGUAAUUUAUGUUUGACACUU-GCGUUGAUUUUUAUUGG--GC-AACACGAUUUGAAGAGCCUGCGGAAAUGUUGCACAUUUCCGUUUCUAGUU-------GCUGCU-CCAUAAC .......(((((.(..((....-..))..)........((--((-(...(((((.(((......(((((((((.....)))))))))))).))))-------).))))-)))))). ( -23.30, z-score = -0.41, R) >consensus UCGUUAUUUAGAUUUGACACUUGGCGCUGAUUUUUAUUGG__GC___CGUUUUUCGGGUAAGCAGCGGAAAUGUUGCACAUUUCCGUUCCAG_________AGCAGCG_CCAUAAC .......................................................(((......(((((((((.....)))))))))))).......................... (-12.10 = -11.57 + -0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:20:19 2011