
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,423,428 – 12,423,520 |
| Length | 92 |
| Max. P | 0.816891 |

| Location | 12,423,428 – 12,423,520 |
|---|---|
| Length | 92 |
| Sequences | 13 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 58.53 |
| Shannon entropy | 0.92208 |
| G+C content | 0.49427 |
| Mean single sequence MFE | -24.33 |
| Consensus MFE | -8.65 |
| Energy contribution | -8.05 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.60 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.816891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12423428 92 - 27905053 ---------GUAGUCCCAGUUUGUCAAUUCAAUAAUACUUU--AUGGGUUUCAGCAACUGGGAGCUCCGAUGCAGGGCACCCACCUGCGAUUGCAGGCCAACC ---------....((((((((.((.((((((.(((....))--)))))))...))))))))))(((.(((((((((.......))))).))))..)))..... ( -27.90, z-score = -1.17, R) >anoGam1.chr2R 2025318 96 + 62725911 -UCAGCUGUACCUUUUCCACUAACUAUUGCACCAUCUCUA---UCAUUUGACAGCAACUCGGUAUUCCUAUGAAAG---CAAACAUUCGUGUACAGAUCAGCC -....((((((...............(((((((....((.---((....)).))......))).(((....))).)---)))((....))))))))....... ( -10.90, z-score = 0.80, R) >droGri2.scaffold_14830 155532 95 + 6267026 -----AUAGAAUAUCUCUCUUCUCUUGAAAUCUAACCAGACGUAAUAUUUACAGCAACUGGGUGUGCCUUUGCAUG---CUCAUAUGCGUCUACAGGCCAACC -----.((((...((...........))..))))...(((((((.....))).(((..(((((((((....)))))---))))..)))))))........... ( -19.60, z-score = -0.94, R) >droMoj3.scaffold_6540 7902710 93 + 34148556 ------UCCGCUUUCUUUGCUUUCUAUGAAGCUCACUUAGC-CAAUCUCUACAGCAACUGGGCGUACCCUUGCACG---UCCAUGUGCGUCUGCAGGCCAACC ------...(((......(((((....)))))......)))-...........(((..(((((((........)))---))))..)))(((....)))..... ( -21.80, z-score = -0.50, R) >droVir3.scaffold_13047 13844420 92 - 19223366 ------GUAAUUUUUACUUGUUUCUUCAAAACUAAC-CGCC-CAAUCCCUACAGCAACUGGGCGUGCCCUUGCAUG---UUCACAUGCGUCUACAGGCCAACC ------..........(((((..(............-((((-((..............)))))).......(((((---....))))))...)))))...... ( -18.94, z-score = -1.23, R) >droWil1.scaffold_181108 107954 101 + 4707319 UCCAUCCGCAUUGGAGGAGCUCUCAUUCUAAUUAAUUCUUC--AUUUUUUGCAGCAAAUUGGAGCUCCAAUGCAAGGAACUCAUUUGCGUCUACAGGCAAAUC ....((((((((((((((....)).((((((((....((.(--(.....)).))..)))))))))))))))))..)))....((((((........)))))). ( -26.00, z-score = -1.92, R) >droPer1.super_19 842497 103 + 1869541 ACCUUUCUUUCUGCGCCGUCUGACUGCAACCAUUUUCCCUUCCUUCCAUUCCAGCAACUGGGAGCUCCCAUGCAGGGCACACAUCUGCGACUGCAGGCCAAUC .........((((((..(((...(((((......(((((....................)))))......))))).(((......))))))))))))...... ( -22.65, z-score = 0.23, R) >dp4.chr2 25478892 103 + 30794189 ACCUUUCUUUCUGCGCAGUCUGACUGCAACAAUUUUCCCUUCCUUCCCUUCCAGCAACUGGGAGCUCCCAUGCAGGGCACACAUCUGCGACUGCAGGCCAAUC ..........((((((((.....))))....................(((((((...))))))).......))))(((......((((....))))))).... ( -26.50, z-score = -0.86, R) >droAna3.scaffold_13340 16233069 96 + 23697760 ----UUUAUCCUUUUUAAACUAACUAAAUAAAUUAUAAAUG--AU-UUCUUUAGCACCUUGGAGCCCCCAUGCACGGCACCCACCUGCGAUUACAGGCCAAUC ----...................(((((.((((((....))--))-)).)))))....((((.(((.........))).....((((......)))))))).. ( -14.70, z-score = -1.05, R) >droEre2.scaffold_4770 8532505 92 + 17746568 ---------GUUGUCCAGGCUGGUCAAUGCAAUAAUACUGU--ACGGGUUUCAGCAAUUGGGAGCUCCGAUGCAGGGCACCCACCUGCGCUUGCAGGCCAACC ---------........((.(((((..(((((.........--.((((((((........))))).)))..(((((.......)))))..)))))))))).)) ( -30.20, z-score = 0.05, R) >droYak2.chr3R 16862687 92 + 28832112 ---------GUAGUCCAGGUUGGUCAAUGCAAUAAUACUUC--GUGGGUUUCAGCAAUUGGGAGCUCCGAUGCAGGGCACCCACCUGCGCUUGCAGGCCAACC ---------........((((((((..(((((.......((--(.(((((((........)))))))))).(((((.......)))))..))))))))))))) ( -35.80, z-score = -2.10, R) >droSec1.super_12 398302 92 + 2123299 ---------GUGGUCCCAGUUGGUCAAUUCAAUAAUACUUU--AUGGGUUUCAGCAACUUGGAGCUCCGAUGCAGGGCACCCACCUGCGAUUGCAGGCCAACC ---------.........(((((((................--..(((((((((....)))))))))(((((((((.......))))).))))..))))))). ( -28.60, z-score = -0.81, R) >droSim1.chr3R 18473573 92 + 27517382 ---------GUGGUCCCAGUUGGUCAAUUCAAUAAUACUUU--AUGGAUUUCAGCAACUGGGAGCUCCGAUGCAGGGCACCCACCUGCGAUUGCAGGCCAACC ---------(((((((((((((.(.((((((.(((....))--)))))))..).)))))))))((.((......))))..))))((((....))))....... ( -32.70, z-score = -2.11, R) >consensus _________GUUGUCCCAGCUGGCUAAUACAAUAAUACUUU__AUGGUUUUCAGCAACUGGGAGCUCCCAUGCAGGGCACCCACCUGCGACUGCAGGCCAACC .....................................................((.....((....))..((((((.......)))))).......))..... ( -8.65 = -8.05 + -0.61)

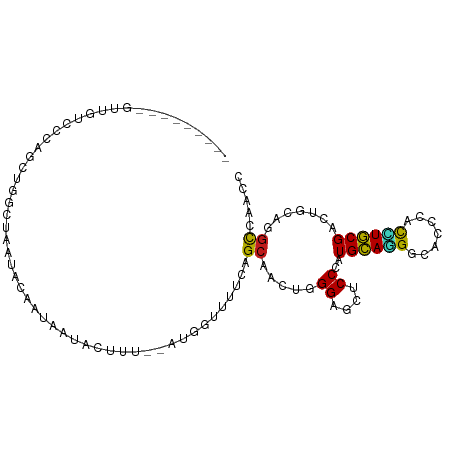
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:20:11 2011