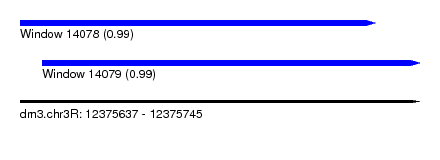
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,375,637 – 12,375,745 |
| Length | 108 |
| Max. P | 0.993727 |

| Location | 12,375,637 – 12,375,733 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.53 |
| Shannon entropy | 0.35605 |
| G+C content | 0.48534 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -25.30 |
| Energy contribution | -25.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.993727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12375637 96 + 27905053 --------------CAUUCGCCAGCAGCUUCUCAGC-CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAACAAC-- --------------..((((..(((.(((....)))-.((((((((......))))))))...............)))...))))(((((((.....))))))).......-- ( -27.70, z-score = -2.19, R) >droSim1.chr3R 18425768 95 - 27517382 ---------------CAGCAGCAGCAGCUUCUCAGC-CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAAUUGCGCAAAAAACAAC-- ---------------.(((.((((((..((.(((((-.....)).))).))..))))))((((((...)))))).))).......(((((((.....))))))).......-- ( -28.10, z-score = -1.74, R) >droSec1.super_12 350995 95 - 2123299 ---------------CAGCAGCAGCAGCUUCUCAGC-CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAACAAC-- ---------------.(((.((((((..((.(((((-.....)).))).))..))))))((((((...)))))).))).......(((((((.....))))))).......-- ( -28.10, z-score = -1.70, R) >droYak2.chr3R 16814427 103 - 28832112 -------CAUUCGCUAUUCGCCAGCAGCUUCUCAGC-CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAACAAC-- -------..((((((.......)))((((.......-..(((((((......)))))))((((((...))))))))))....)))(((((((.....))))))).......-- ( -28.30, z-score = -1.79, R) >droEre2.scaffold_4770 8484529 103 - 17746568 -------CACUAGCUAUACGCCAGCAGAUUCUCAGC-CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAAGAAC-- -------.....((.........((((((((..(((-..(((((((......)))))))((((((...)))))).)))....))))))))((.....))))..........-- ( -28.70, z-score = -2.17, R) >droAna3.scaffold_13340 16186755 95 - 23697760 ------------------UGGGAGCUUCUUCUCUCCGCCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAACAACCC ------------------..((((........))))..((((((((......)))))))).....((((((.......)))))).(((((((.....)))))))......... ( -29.70, z-score = -2.96, R) >dp4.chr2 25429534 111 - 30794189 UGUCUGCCCCAAAGCCCCCUGCGCCCCCCCUCCGUCCCCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAACAAC-- .............((.....))................((((((((......)))))))).....((((((.......)))))).(((((((.....))))))).......-- ( -25.80, z-score = -2.95, R) >droPer1.super_19 790720 111 - 1869541 UGUCUGCCCCAAAGCCCCCUGCGCCCCCCCUCCGUCCCCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAACAAC-- .............((.....))................((((((((......)))))))).....((((((.......)))))).(((((((.....))))))).......-- ( -25.80, z-score = -2.95, R) >consensus _______________CUCCGGCAGCAGCUUCUCAGC_CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAACAAC__ ......................................((((((((......)))))))).....((((((.......)))))).(((((((.....)))))))......... (-25.30 = -25.30 + 0.00)
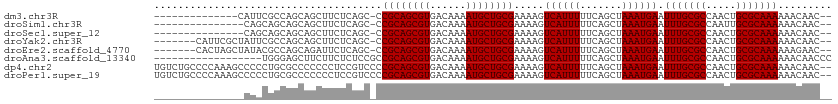
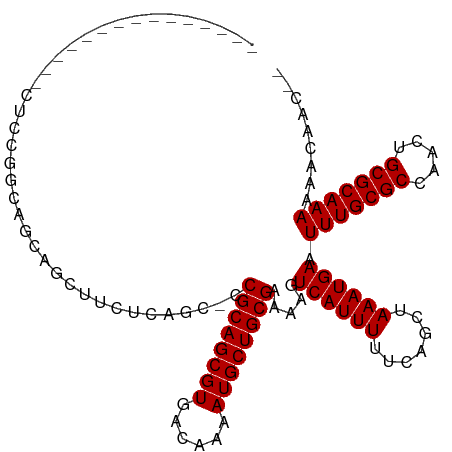
| Location | 12,375,643 – 12,375,745 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 86.80 |
| Shannon entropy | 0.24765 |
| G+C content | 0.46604 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -25.30 |
| Energy contribution | -25.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.993206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12375643 102 + 27905053 -------CCAGCAGCUUCUCAGC----CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAACAACGC--CCAAAAAAGU -------...((((((.......----..(((((((......)))))))((((((...)))))))))).......(((((((.....))))))).......))--.......... ( -27.40, z-score = -2.07, R) >droSim1.chr3R 18425774 101 - 27517382 --------CAGCAGCUUCUCAGC----CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAAUUGCGCAAAAAACAACGC--CCAAAAAAGU --------..((((((.......----..(((((((......)))))))((((((...)))))))))).......(((((((.....))))))).......))--.......... ( -27.40, z-score = -2.00, R) >droSec1.super_12 351001 101 - 2123299 --------CAGCAGCUUCUCAGC----CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAACAACGC--CCAAAAAAGU --------..((((((.......----..(((((((......)))))))((((((...)))))))))).......(((((((.....))))))).......))--.......... ( -27.40, z-score = -1.98, R) >droYak2.chr3R 16814435 107 - 28832112 --AUUCGCCAGCAGCUUCUCAGC----CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAACAACGC--CCAAAAAAGU --....((.(((.(((....)))----.((((((((......))))))))...............))).......(((((((.....))))))).......))--.......... ( -27.80, z-score = -1.59, R) >droEre2.scaffold_4770 8484537 107 - 17746568 --AUACGCCAGCAGAUUCUCAGC----CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAAGAACGC--CCAAAAAAGU --....((..((((((((..(((----..(((((((......)))))))((((((...)))))).)))....))))))))((.....))))............--.......... ( -28.40, z-score = -2.09, R) >droAna3.scaffold_13340 16186758 104 - 23697760 --------GAGCUUCUUCUCUCCG---CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAACAACCCGACGAAAAAAAA --------(((......)))....---.((((((((......))))))))....(((....((((......))))(((((((.....))))))).........)))......... ( -28.60, z-score = -3.01, R) >dp4.chr2 25429544 113 - 30794189 AAAGCCCCCUGCGCCCCCCCUCCGUCCCCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAACAACGA--CCAAGAAGAU ...((.....)).......((..(((..((((((((......)))))))).....((((((.......)))))).(((((((.....))))))).......))--)..))..... ( -28.20, z-score = -3.70, R) >droPer1.super_19 790730 113 - 1869541 AAAGCCCCCUGCGCCCCCCCUCCGUCCCCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAACAACGA--CCAAGAAAAU ...((.....)).......((..(((..((((((((......)))))))).....((((((.......)))))).(((((((.....))))))).......))--)..))..... ( -28.20, z-score = -3.98, R) >consensus _______CCAGCAGCUUCUCAGC____CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAACAACGC__CCAAAAAAGU ............................((((((((......)))))))).....((((((.......)))))).(((((((.....)))))))..................... (-25.30 = -25.30 + 0.00)

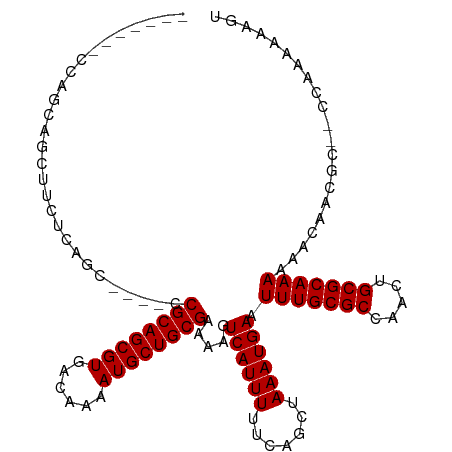
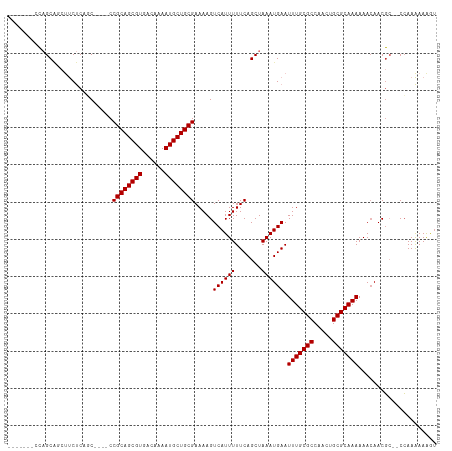
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:20:07 2011