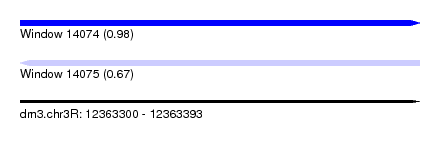
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,363,300 – 12,363,393 |
| Length | 93 |
| Max. P | 0.976642 |

| Location | 12,363,300 – 12,363,393 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 72.82 |
| Shannon entropy | 0.54802 |
| G+C content | 0.46735 |
| Mean single sequence MFE | -19.32 |
| Consensus MFE | -13.73 |
| Energy contribution | -13.98 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.976642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
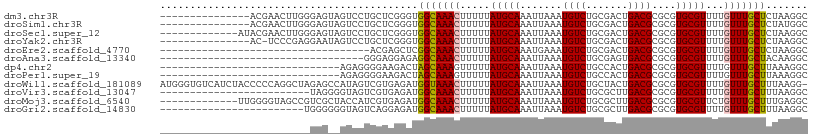
>dm3.chr3R 12363300 93 + 27905053 GCCUUAGAGCAAACAAAACGCACGCGCGUCAGUCGCAGACAUUUAAUUUGCAUAAAAAGUUUGCCACCCGAGCAGGACUACUCCCAAGUUCGU--------------- ......(.((((((...((((....)))).....(((((.......))))).......)))))))...(((((.(((....)))...))))).--------------- ( -22.00, z-score = -1.65, R) >droSim1.chr3R 18413431 93 - 27517382 GCCAUAGAGCAAACAAAACGCACGCGCGUCAGUCGCAGACAUUUAAUUUGCAUAAAAAGUUUGCCACCCGAGCAGGACUACUCCCAAGUUCGU--------------- ......(.((((((...((((....)))).....(((((.......))))).......)))))))...(((((.(((....)))...))))).--------------- ( -22.00, z-score = -1.91, R) >droSec1.super_12 338720 95 - 2123299 GCCUUAGAGCAAACAAAACGCACGCGCGUCAGUCGCAGACAUUUAAUUUGCAUAAAAAGUUUGCCACCCGAGCAGGACUACUCCCAAGUUCGUAU------------- ......(.((((((...((((....)))).....(((((.......))))).......)))))))...(((((.(((....)))...)))))...------------- ( -22.00, z-score = -1.72, R) >droYak2.chr3R 16801737 92 - 28832112 GCCUUAGAGCAAACAAAACGCACGCGCGUCAGUCGCAGACAUUUAAUUUGCAUAAAAAGUUUGCCACCCGAGCAGGACUAUUCCUCGGGA-GU--------------- ......(.((((((...((((....)))).....(((((.......))))).......))))))).((((((.((.....)).)))))).-..--------------- ( -25.50, z-score = -1.95, R) >droEre2.scaffold_4770 8472171 73 - 17746568 GCCUUAGAGCAAACAAAACGCACGCGCGUCAGUCGCAGACAUUUCAUUUGCAUAAAAAGUUUGCCGAGCUCGU----------------------------------- ......((((.......((((....))))...((((((((.(((.((....)).))).))))).)))))))..----------------------------------- ( -16.60, z-score = -0.29, R) >droAna3.scaffold_13340 16175062 74 - 23697760 GCCUUGUAGCAAACAAAACGCACGCGCGUCACUCGCAGACAUUUAAUUUGCAUAAAAAGUUUGCCUCUCCUCCC---------------------------------- .....(..((((((...((((....)))).....(((((.......))))).......))))))..).......---------------------------------- ( -15.60, z-score = -2.15, R) >dp4.chr2 25419383 78 - 30794189 GCCUUUAAGCAAACAAAACGCACGCGCGUCAGUGGCAGACAUUUAAUUUGCAUAAAAACUUUGCUAGUCUUCCCCUCU------------------------------ .......((((((((..((((....))))...))(((((.......)))))........)))))).............------------------------------ ( -13.90, z-score = -0.43, R) >droPer1.super_19 780573 78 - 1869541 GCCUUUAAGCAAACAAAACGCACGCGCGUCAGUGGCAGACAUUUAAUUUGCAUAAAAACUUUGCUAGUCUUCCCCUCU------------------------------ .......((((((((..((((....))))...))(((((.......)))))........)))))).............------------------------------ ( -13.90, z-score = -0.43, R) >droWil1.scaffold_181089 7092911 107 - 12369635 -CCUUAAAGCAAACAAAACGCACGCGCGUCAGUAGCAGACAUUUAAUUUGCAUAAAAAGUUUACCAUCUCACGACUAUGGCUCUAGCCUGGGGGUAGAUGACACCCAU -.........((((...((((....)))).....(((((.......))))).......))))..(((((.((..(((.(((....))))))..)))))))........ ( -23.00, z-score = -0.52, R) >droVir3.scaffold_13047 13780017 83 + 19223366 GCCUUAAAGCAAACAAAACGCACGCGCGUCAAGCGCAGACAUUUAAUUUGCAUAAAAAGUUUGCCAUCUCACGACUACCCCUA------------------------- ........((((((...((((....)))).....(((((.......))))).......))))))...................------------------------- ( -15.20, z-score = -1.34, R) >droMoj3.scaffold_6540 7835807 95 - 34148556 GCCUCAAAGCAAACAGAACGCACGCGCGUCAAGCGCAGACAUUUAAUUUGCAUAAAAAGUUUGCCAUCUCACGAUGGUAGCGACGGCUACCCCAA------------- (((((...(((((..(((.(...((((.....))))...).)))..))))).......(((.((((((....))))))))))).)))........------------- ( -27.00, z-score = -2.19, R) >droGri2.scaffold_14830 96232 84 - 6267026 GCCUUAAAGCAAACAAAACGCACGCGCGUCAAGCGCAGACAUUUAAUUUGCAUAAAAAGUUUGCCAUCUCCUGACUACCCCCCA------------------------ ........((((((...((((....)))).....(((((.......))))).......))))))....................------------------------ ( -15.20, z-score = -1.67, R) >consensus GCCUUAAAGCAAACAAAACGCACGCGCGUCAGUCGCAGACAUUUAAUUUGCAUAAAAAGUUUGCCACCCCACCACGAC__CUC_________________________ ........((((((...((((....)))).....(((((.......))))).......))))))............................................ (-13.73 = -13.98 + 0.25)



| Location | 12,363,300 – 12,363,393 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 72.82 |
| Shannon entropy | 0.54802 |
| G+C content | 0.46735 |
| Mean single sequence MFE | -25.05 |
| Consensus MFE | -14.23 |
| Energy contribution | -14.18 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.670750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12363300 93 - 27905053 ---------------ACGAACUUGGGAGUAGUCCUGCUCGGGUGGCAAACUUUUUAUGCAAAUUAAAUGUCUGCGACUGACGCGCGUGCGUUUUGUUUGCUCUAAGGC ---------------.....((((((((((....)))))((((.....)))).....((((((.((((((.((((.......)))).)))))).)))))).))))).. ( -26.30, z-score = -0.54, R) >droSim1.chr3R 18413431 93 + 27517382 ---------------ACGAACUUGGGAGUAGUCCUGCUCGGGUGGCAAACUUUUUAUGCAAAUUAAAUGUCUGCGACUGACGCGCGUGCGUUUUGUUUGCUCUAUGGC ---------------..(.(((((((((.....)).)))))))..)...........((((((.((((((.((((.......)))).)))))).))))))........ ( -22.90, z-score = 0.48, R) >droSec1.super_12 338720 95 + 2123299 -------------AUACGAACUUGGGAGUAGUCCUGCUCGGGUGGCAAACUUUUUAUGCAAAUUAAAUGUCUGCGACUGACGCGCGUGCGUUUUGUUUGCUCUAAGGC -------------.......((((((((((....)))))((((.....)))).....((((((.((((((.((((.......)))).)))))).)))))).))))).. ( -26.30, z-score = -0.59, R) >droYak2.chr3R 16801737 92 + 28832112 ---------------AC-UCCCGAGGAAUAGUCCUGCUCGGGUGGCAAACUUUUUAUGCAAAUUAAAUGUCUGCGACUGACGCGCGUGCGUUUUGUUUGCUCUAAGGC ---------------.(-.((((((.(.......).)))))).).....((((....((((((.((((((.((((.......)))).)))))).))))))...)))). ( -28.70, z-score = -1.62, R) >droEre2.scaffold_4770 8472171 73 + 17746568 -----------------------------------ACGAGCUCGGCAAACUUUUUAUGCAAAUGAAAUGUCUGCGACUGACGCGCGUGCGUUUUGUUUGCUCUAAGGC -----------------------------------.((....)).....((((....(((((..((((((.((((.......)))).))))))..)))))...)))). ( -21.00, z-score = -0.74, R) >droAna3.scaffold_13340 16175062 74 + 23697760 ----------------------------------GGGAGGAGAGGCAAACUUUUUAUGCAAAUUAAAUGUCUGCGAGUGACGCGCGUGCGUUUUGUUUGCUACAAGGC ----------------------------------....((((((.....))))))..((((((.((((((.((((.......)))).)))))).))))))........ ( -19.30, z-score = -0.34, R) >dp4.chr2 25419383 78 + 30794189 ------------------------------AGAGGGGAAGACUAGCAAAGUUUUUAUGCAAAUUAAAUGUCUGCCACUGACGCGCGUGCGUUUUGUUUGCUUAAAGGC ------------------------------.(..((.....))..)...(((((((.((((((.((((((.((((......).))).)))))).)))))).))))))) ( -19.70, z-score = -0.25, R) >droPer1.super_19 780573 78 + 1869541 ------------------------------AGAGGGGAAGACUAGCAAAGUUUUUAUGCAAAUUAAAUGUCUGCCACUGACGCGCGUGCGUUUUGUUUGCUUAAAGGC ------------------------------.(..((.....))..)...(((((((.((((((.((((((.((((......).))).)))))).)))))).))))))) ( -19.70, z-score = -0.25, R) >droWil1.scaffold_181089 7092911 107 + 12369635 AUGGGUGUCAUCUACCCCCAGGCUAGAGCCAUAGUCGUGAGAUGGUAAACUUUUUAUGCAAAUUAAAUGUCUGCUACUGACGCGCGUGCGUUUUGUUUGCUUUAAGG- ..(((((.....)))))((.(((....)))...(((....)))))....(((.....((((((.((((((.(((.........))).)))))).))))))...))).- ( -29.60, z-score = -1.03, R) >droVir3.scaffold_13047 13780017 83 - 19223366 -------------------------UAGGGGUAGUCGUGAGAUGGCAAACUUUUUAUGCAAAUUAAAUGUCUGCGCUUGACGCGCGUGCGUUUUGUUUGCUUUAAGGC -------------------------.....((.(((....))).))...((((....((((((.((((((.(((((.....))))).)))))).))))))...)))). ( -27.20, z-score = -2.21, R) >droMoj3.scaffold_6540 7835807 95 + 34148556 -------------UUGGGGUAGCCGUCGCUACCAUCGUGAGAUGGCAAACUUUUUAUGCAAAUUAAAUGUCUGCGCUUGACGCGCGUGCGUUCUGUUUGCUUUGAGGC -------------(((.((((((....))))))(((....)))..))).((((....((((((..(((((.(((((.....))))).)))))..))))))...)))). ( -34.10, z-score = -2.21, R) >droGri2.scaffold_14830 96232 84 + 6267026 ------------------------UGGGGGGUAGUCAGGAGAUGGCAAACUUUUUAUGCAAAUUAAAUGUCUGCGCUUGACGCGCGUGCGUUUUGUUUGCUUUAAGGC ------------------------((((((((.((((.....))))..)))))))).((((((.((((((.(((((.....))))).)))))).))))))........ ( -25.80, z-score = -1.75, R) >consensus _________________________GAG__GUAGUGGUGGGGUGGCAAACUUUUUAUGCAAAUUAAAUGUCUGCGACUGACGCGCGUGCGUUUUGUUUGCUUUAAGGC ...........................................(((((((.....(((((.......((((.......))))....)))))...)))))))....... (-14.23 = -14.18 + -0.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:20:04 2011