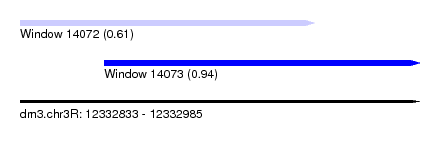
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,332,833 – 12,332,985 |
| Length | 152 |
| Max. P | 0.937023 |

| Location | 12,332,833 – 12,332,945 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 71.48 |
| Shannon entropy | 0.49829 |
| G+C content | 0.41071 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -13.50 |
| Energy contribution | -14.98 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.608753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12332833 112 + 27905053 GGACGGAUUAAAAUGGUCAGCUUAAUUCCCGGAUUAUCGAUACUUUGAGUCUUGGGACUUAUGGGCAGAUUAUCUCUGCCUGAUGAGUAAUGGUUGUUUCGGUUCUUAAGUA ....((((..((((..(((........(((((((..((((....))))))).))))(((((((((((((.....))))))).))))))..)))..))))..))))....... ( -33.90, z-score = -2.17, R) >droSim1.chr3R 18383262 93 - 27517382 UGUAGGAUCAAAAUGGUCAGCUUAAUUCCUGGAUUAUCGAUACUUUAAGUCUUGGGACUUAUGGGCAGAUUAUCUCUGCCUGGUUUUUAAGUA------------------- .....((((.....)))).((((((..((................((((((....)))))).(((((((.....)))))))))...)))))).------------------- ( -25.60, z-score = -1.93, R) >droSec1.super_12 308699 85 - 2123299 UGUCGGAUCAAAAUGGUCAGUUUAAUUCCUGGAUUAACGAUACUUUAAGUCUUGGGAC--------AGAUUAUCUCUGCCUGGUUUUUAAGUA------------------- (((((((((.....))))...((((((....))))))))))).((((((..(..((.(--------(((.....))))))..)..))))))..------------------- ( -20.20, z-score = -1.32, R) >droYak2.chr3R 16770459 112 - 28832112 UGUCGGAUCAAAGCGGUCACCUUGGUUCCGGGAUAAUCAAUACUUUGAGCCUUGGGACUUAUGGGCAGAUUAUCUCUGCCUGAAGGGUAAUGUAAGCUUCGGAUUUUAAACG ....(((((.((((.((.((((((((((((((....((((....))))..)))))))))...(((((((.....)))))))..))))).))....))))..)))))...... ( -33.10, z-score = -1.34, R) >droEre2.scaffold_4770 8441017 112 - 17746568 UGUCAGAUCAAAGCGGUCAGCUUAAUUCCGGGAUUGUCAAUACUUUGGGUCUGGGGACUUAUGGCCAGAUUAUCGCUGCCUGAUGAGUAAUGGCUAUUUCGGAUUUUAAACG .....((((.....)))).(.((((.((((..((.((((.(((((((((((....)))))).(((.((.......)))))....))))).)))).))..))))..)))).). ( -30.80, z-score = -0.83, R) >consensus UGUCGGAUCAAAAUGGUCAGCUUAAUUCCGGGAUUAUCGAUACUUUGAGUCUUGGGACUUAUGGGCAGAUUAUCUCUGCCUGAUGAGUAAUGA____UUCGG_U_UUAA___ .....((((.....)))).......((((.((((..((((....)))))))).)))).....(((((((.....)))))))............................... (-13.50 = -14.98 + 1.48)


| Location | 12,332,865 – 12,332,985 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.53 |
| Shannon entropy | 0.46850 |
| G+C content | 0.33136 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -14.22 |
| Energy contribution | -16.42 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.937023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12332865 120 + 27905053 AUUAUCGAUACUUUGAGUCUUGGGACUUAUGGGCAGAUUAUCUCUGCCUGAUGAGUAAUGGUUGUUUCGGUUCUUAAGUAAAUAAUUUCUUGAGAAAUUUGUUUGAGAAUAUGAGGUAAU .(((((.......((((((....)))))).(((((((.....))))))))))))......((((..((((((((((((.....((((((....))))))..))))))))).)))..)))) ( -32.80, z-score = -2.97, R) >droSim1.chr3R 18383294 101 - 27517382 AUUAUCGAUACUUUAAGUCUUGGGACUUAUGGGCAGAUUAUCUCUGCCUG-------------------GUUUUUAAGUAAAUCAUUUCUUGGGAAAUUUGUUGUAGAAUAUGAGGUAAU ........(((((((..(((..(((.....(((((((.....)))))))(-------------------((((......)))))...)))..))).((((......)))).))))))).. ( -24.60, z-score = -1.97, R) >droSec1.super_12 308731 93 - 2123299 AUUAACGAUACUUUAAGUCUUGGGAC--------AGAUUAUCUCUGCCUG-------------------GUUUUUAAGUAAAUCAUUUCUUGAGAAAUUUGUUUGAGAAUAUGAGGUAAU ........(((((((..(((..(.((--------(((((.((((....((-------------------((((......))))))......))))))))))))..)))...))))))).. ( -22.30, z-score = -2.17, R) >droYak2.chr3R 16770491 115 - 28832112 AUAAUCAAUACUUUGAGCCUUGGGACUUAUGGGCAGAUUAUCUCUGCCUGAAGGGUAAUGUAAGCUUCGGAUUUUAAACGA-----UUCUUGAGAAAUUUGUUUGAAAGAACAAAAUAAU ....((((....))))..((..((((((.((((((((.....)))))))).)))............(((.........)))-----.)))..))...(((((((....)))))))..... ( -27.80, z-score = -2.04, R) >droEre2.scaffold_4770 8441049 115 - 17746568 AUUGUCAAUACUUUGGGUCUGGGGACUUAUGGCCAGAUUAUCGCUGCCUGAUGAGUAAUGGCUAUUUCGGAUUUUAAACGA-----UUCUUGAGAAAUUUGUUUGAAAGAAAAGAAUAAU .............((((((....))))))((((((..((((((.....))))))....)))))).(((...((((((((((-----(((....)))..)))))))))).....))).... ( -24.80, z-score = -0.67, R) >consensus AUUAUCGAUACUUUGAGUCUUGGGACUUAUGGGCAGAUUAUCUCUGCCUGA_G_GUAAUG_____UUCGGUUUUUAAGUAAAU_AUUUCUUGAGAAAUUUGUUUGAGAAUAUGAGGUAAU ........(((((((((((....)))))..(((((((.....)))))))....................((((((((((((((...(((....)))))))))))))))))..)))))).. (-14.22 = -16.42 + 2.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:20:03 2011