| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,320,382 – 12,320,472 |
| Length | 90 |
| Max. P | 0.778666 |

| Location | 12,320,382 – 12,320,472 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 62.67 |
| Shannon entropy | 0.71767 |
| G+C content | 0.55027 |
| Mean single sequence MFE | -26.91 |
| Consensus MFE | -8.19 |
| Energy contribution | -7.91 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.778666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
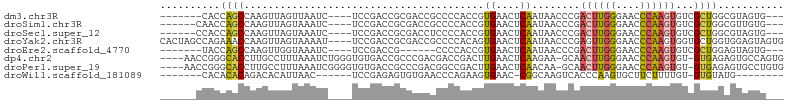
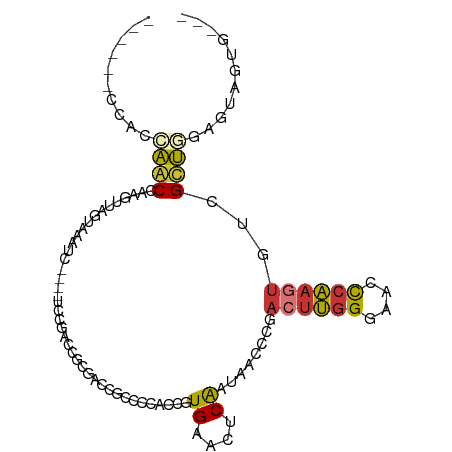
>dm3.chr3R 12320382 90 - 27905053 -------CACCAGCCAAGUUAGUUAAUC----UCCGACCGCGACCGCCCCACCGUGAACUCAAUAACCCGACUUGGGAACCCAAGUGUCGCUGGCGUAGUG--- -------(((..((((.((..(......----.)..)).((((((((......)))..............((((((....)))))))))))))))...)))--- ( -24.80, z-score = -1.68, R) >droSim1.chr3R 18370788 91 + 27517382 ------CAACCAGCCAAGUUAGUAAAUC----UCCGACCGCGACCGCCCCACCGUGAACUCAAUAACCCGACUUGGGAACCCAAGUGUCGCUGGCGUUGUG--- ------((((..((((.((..(......----.)..)).((((((((......)))..............((((((....)))))))))))))))))))..--- ( -24.70, z-score = -1.65, R) >droSec1.super_12 296192 91 + 2123299 ------CCACCAGCCAAGUUAGUAAAUC----UCCGACCGCGACCUCCCCACCGUGAACUCAAUAACCCGACUUGGGAACCCAAGUGUCGCUGGCGUAGUG--- ------.(((..((((.((..(......----.)..)).(((((.........(....)...........((((((....)))))))))))))))...)))--- ( -23.60, z-score = -1.78, R) >droYak2.chr3R 16757989 100 + 28832112 CACUAGCCAGAAACCAAGUUAGUAAAAU----UCCGACCGCGACCGCCCCACAGUGAACUCAAUAACCCGAGUUGGGAACCCAAGUGGUGCUGGUGGAGUAGUG .((((((..(....)..))))))...((----(((.(((((.(((((.........(((((........)))))((....))..))))))).)))))))).... ( -28.40, z-score = -1.72, R) >droEre2.scaffold_4770 8425224 84 + 17746568 -------UACCAGCCAAGUUGGUAAAUC----UCCGACCG------CCCCACCGUGAACUCAAUAACCCGACUUGGGAACCCAAGUGUCGCUGGAGUAGUG--- -------..(((((...(((((......----.)))))((------(......)))..............((((((....))))))...))))).......--- ( -22.70, z-score = -1.51, R) >dp4.chr2 25381264 98 + 30794189 ----AACCGGGCACCUUGCCUUUAAAUCUGGGUGUGACCGCCCGACGACCGACUUGAACUCAAGAA-GCAACUUGGGAACCCAAGUGU-GUGAGAGUGCCAGUG ----.....(((((((..(.........((((((....))))))........((((....))))..-(((.(((((....))))))))-)..)).))))).... ( -34.10, z-score = -2.32, R) >droPer1.super_19 740284 98 + 1869541 ----AACCGGGCACCUUGCCUUUAAAUCGGGGUGUGACCGCCCGACGGCCGACUUGAACUCAACAA-GCAACUUGGGAACCCAAGUGU-GUGAGAGUGCCUGUG ----...(((((((((..(.(((((.(((((......))(((....)))))).)))))........-(((.(((((....))))))))-)..)).))))))).. ( -35.70, z-score = -1.72, R) >droWil1.scaffold_181089 7050029 81 + 12369635 -------CACACACAGACACAUUAAC------UCCGAGAGUGUGAACCCAGAAGUGAAC-CGGCAAGUCACCCAAGUGCUUCUUUUGU-GUGUAUG-------- -------.(((((((((((((((...------......)))))).....(((((((...-.((.......))....))))))))))))-))))...-------- ( -21.30, z-score = -1.35, R) >consensus ______CCACCAACCAAGUUAGUAAAUC____UCCGACCGCGACCGCCCCACCGUGAACUCAAUAACCCGACUUGGGAACCCAAGUGUCGCUGGAGUAGUG___ ..........((((........................................((....))........((((((....))))))...))))........... ( -8.19 = -7.91 + -0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:19:58 2011