
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,297,684 – 12,297,835 |
| Length | 151 |
| Max. P | 0.592775 |

| Location | 12,297,684 – 12,297,835 |
|---|---|
| Length | 151 |
| Sequences | 7 |
| Columns | 152 |
| Reading direction | reverse |
| Mean pairwise identity | 71.25 |
| Shannon entropy | 0.54182 |
| G+C content | 0.50957 |
| Mean single sequence MFE | -38.91 |
| Consensus MFE | -21.85 |
| Energy contribution | -23.35 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.592775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12297684 151 - 27905053 UCCCCCCUUUUUUUUUUUUGAUACUUCACAGCGGCCAUAA-ACGUGACAGCGUAUUUAAUUAGCGCCCUUCGCAAGCGGCCCCCUUCCUGUCCCUUCAAAUUGAUUGGGCCACGUGCUGGAAGGGCGUUGACAUCUCACUUGUAGCUCUCGC ..................(((....)))..(((((.((((-((((....))))......(((((((((((((((.(.(((((...((.((......))....))..))))).).)))..))))))))))))........)))).))...))) ( -40.30, z-score = -0.94, R) >droSim1.chr3R 18352018 145 + 27517382 ------UUUUUUUUUUUUUGGUACCUCACAGCGGCGAUAA-ACGUGACAACGUAUUUAAUUAGCGCCCUUCGCAAGCGGCCCCCUUCUUGUCCCUUCAAAUUGAUUGGGCCACGUGCUGGAAGGGCGUUGACAUCUCACUUGUAGCUCUCGC ------.......................(((.((((...-((((....))))......(((((((((((((((.(.(((((...(((((......)))...))..))))).).)))..))))))))))))........)))).)))..... ( -42.50, z-score = -1.47, R) >droSec1.super_12 273577 140 + 2123299 -----------UUUUUUUUGGUACCUCACAGCGGCGAUAA-ACGUGACAACGUAUUUAAUUAGCGCCCUUCGCAAGCGGCCCCCUUCUUGUCCCUUCAAAUUGAUUGGGCCACGUGCUGGAAGGGCGUUGACAUCUCACUUGUAGCUCUCGC -----------..................(((.((((...-((((....))))......(((((((((((((((.(.(((((...(((((......)))...))..))))).).)))..))))))))))))........)))).)))..... ( -42.50, z-score = -1.41, R) >droYak2.chr3R 14716077 137 + 28832112 -----------UUUUUUUGUGUACCUCACAGCGGCCAUAA-ACGUGACAAUGUAUUUAAUUAGCGCCCUUCUCAAGCGGCCCCCU---UGUCCAUCCAAAUCGAAUGGGCCACGUGCUGGAAGGGCGUUGACAUCUCACUUGUGGCUUUCGC -----------......((((.....))))((((((((((-..((((..((((.......((((((((((((((.(.(((((..(---((...........)))..))))).).))..)))))))))))))))).)))))))))))...))) ( -48.31, z-score = -3.27, R) >droEre2.scaffold_4770 11829474 135 - 17746568 -------------UUUUUUGUUACCUCACAGCGGCCAUAA-ACGUGACAAAGUAUUUAAUUAGCGCCCUUCUCAAGCGGCCCCCU---UGUCCGUCUAAAUCGAAUGGGCCACGUGCUGGAAGGGCGUUGACAUCUCACUUGUGGCUUUCGC -------------.................((((((((((-..((((............(((((((((((((((.(.(((((..(---((...........)))..))))).).))..)))))))))))))....)))))))))))...))) ( -43.29, z-score = -2.05, R) >droAna3.scaffold_13340 15649234 119 - 23697760 ----------------------------UUCUGGUUGUAA-ACGAGACGACGUAUUUAAUUAGUGCUCUGCCCCACCAACCCCCA----AGACAACCAAAUUGAUUGGCCCACGUGCAGGGAGGCCGUUGACAUCUCACUUGUGGCUCAUGC ----------------------------....(((..(((-..(((((((((((((.....)))))...(((((..((....(((----(..(((.....))).))))......))...)).))).))))...))))..)))..)))..... ( -24.40, z-score = 1.77, R) >droVir3.scaffold_12855 4357423 124 + 10161210 --------------------------CACACAGGGCCUGGCGCGUGACAGCGCACUUAAUUAGGAGCCCCCCCUAUUACCUCGCUCCCCGCUGGAUGGCAUCGAUUCGGCAAACAAUUCGGAA--CGUUAAUAUUUCACUUGUGGCUUAUGC --------------------------..(((((((((.(((((((....)))).........(((((...............)))))..)))....)))..((.(((((........))))).--))...........))))))........ ( -31.06, z-score = 1.09, R) >consensus ___________UUUUUUUUGGUACCUCACAGCGGCCAUAA_ACGUGACAACGUAUUUAAUUAGCGCCCUUCGCAAGCGGCCCCCU_C_UGUCCCUUCAAAUUGAUUGGGCCACGUGCUGGAAGGGCGUUGACAUCUCACUUGUGGCUCUCGC ...........................................((.((((.((......((((((((((((...((((((((........................)))))....))).))))))))))))......)))))).))...... (-21.85 = -23.35 + 1.50)
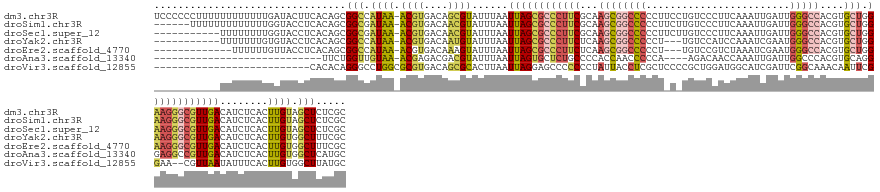

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:19:58 2011