
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,293,773 – 12,293,869 |
| Length | 96 |
| Max. P | 0.573826 |

| Location | 12,293,773 – 12,293,869 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 69.12 |
| Shannon entropy | 0.63334 |
| G+C content | 0.50330 |
| Mean single sequence MFE | -30.53 |
| Consensus MFE | -11.52 |
| Energy contribution | -12.41 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.573826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
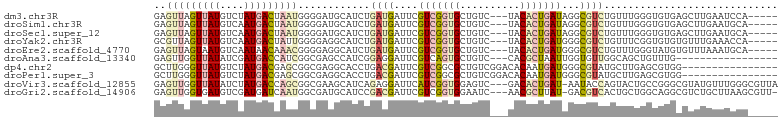
>dm3.chr3R 12293773 96 + 27905053 GAGUUAGUUAUGUCUAUGACUAAUGGGGAUGCAUCUGAUGAUUCGUCGGUGCUGUC---UACACUGAUAGGCGUCUGUUUGGGUGUGAGCUUGAAUCCA----- ..(((((((((....)))))))))((((((((..((((((...))))))..(((((---......)))))))))))(((..(((....)))..))))).----- ( -30.10, z-score = -1.73, R) >droSim1.chr3R 18348113 96 - 27517382 GAGUUAGUUAUGUCAAUGACUAAUGGGGAUGCAUCUGAUGAUUCGUCGGUGCUGUC---UACACUGAUAGGCGUCUGUUUGGGUGUGAGCUUGAAUGCA----- ..(((((((((....)))))))))..((((((..((((((...))))))..(((((---......)))))))))))(((..(((....)))..)))...----- ( -30.10, z-score = -1.80, R) >droSec1.super_12 269676 96 - 2123299 GAGUUAGUUAUGUCAAUGACUAAUGGGGAUGCAUCUGAUGAUUCGUCGGUGCUGUC---UACACUGAUAGGCGUCUGUUUGGGUGUGAGCUUGAAUGCA----- ..(((((((((....)))))))))..((((((..((((((...))))))..(((((---......)))))))))))(((..(((....)))..)))...----- ( -30.10, z-score = -1.80, R) >droYak2.chr3R 14712135 96 - 28832112 GCGUUAGUUAUGUCAAUGACUAUUGGGGAGGCAUCUGAUGAUUCGUCGGUGCUGUC---UACACUGAUGGGCGUCUGUUUCGGUGUGUGUUUGAAACCA----- .((.(((((((....))))))).))..((((((...((((.((((((((((.....---..))))))))))))))))))))(((...........))).----- ( -28.50, z-score = -1.00, R) >droEre2.scaffold_4770 11825428 96 + 17746568 GAGUUAGUAAUGUCAAUAACAAACGGGGAGGCAUCUGAUGAUUCGUCGGUGCUGUC---UACACUGAUGGGCGUCUGUUUGGGUAUGUGUUUAAAUGCA----- ...................((((((((..((((.((((((...))))))))))(((---((......))))).)))))))).((((........)))).----- ( -23.90, z-score = -0.36, R) >droAna3.scaffold_13340 15645634 84 + 23697760 GAGUUGGUUAUAUCGAUGACCAUCGGCGAGCCAUCGGAGGAUUCGUCAGUGCUGUC---CACGCUAAUUGGUGUUGGCAGCUGUUUG----------------- ..(.(((((((....))))))).)((((((((......)).))))))...((((((---.(((((....))))).))))))......----------------- ( -31.20, z-score = -2.25, R) >dp4.chr2 21496970 88 + 30794189 GCUUGGGUUAUGUCUAUGACGAGCGGCGAGGCACCUGACGAUUCGUCGGCGCUGUCGGACACAAUGAUGGGCGUAUGCUUGAGCGUGG---------------- (((..(((((((((((((((.((((.(.((....))((((...))))).)))))))(....)....))))))))).)))..)))....---------------- ( -33.60, z-score = -1.42, R) >droPer1.super_3 4281015 88 + 7375914 GCUUGGGUUAUGUCUAUGACGAGCGGCGAGGCACCUGACGAUUCGUCGGCGCUGUCGGACACAAUGAUGGGCGUAUGCUUGAGCGUGG---------------- (((..(((((((((((((((.((((.(.((....))((((...))))).)))))))(....)....))))))))).)))..)))....---------------- ( -33.60, z-score = -1.42, R) >droVir3.scaffold_12855 5787536 100 + 10161210 GAGUUGGUUAUAUCUAUGACCAGCGGCGAAGCAUCAGAGGAUUCAUCGGUGGAGUC---GACACUGAU-AAUACCAGUACUGCCGGGCGUAUGUUUGGGCGUUA ..(((((((((....)))))))))(((((....))...(((((.(((((((.....---..)))))))-))).))......))).(((((........))))). ( -30.40, z-score = -0.80, R) >droGri2.scaffold_14906 5949515 99 - 14172833 GAGUUGGUGAUGUCGAUGAUCAAUGGCGAUGCAUCCGACGAUUCGUCGGUGGAAUC---AACGCUUAU-GACGUCACUGCUGGCAGGCGUCUGCUUAAGCGUU- .....((((((((((((((((......))).))))(((((...)))))(((.....---..)))....-)))))))))((((((((....)))))..)))...- ( -33.80, z-score = -0.67, R) >consensus GAGUUAGUUAUGUCAAUGACUAACGGCGAGGCAUCUGAUGAUUCGUCGGUGCUGUC___UACACUGAUGGGCGUCUGUUUGGGUGUGAGUUUGAAUGCA_____ ..(((((((((....)))))))))............((((....(((((((..........)))))))...))))............................. (-11.52 = -12.41 + 0.89)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:19:56 2011