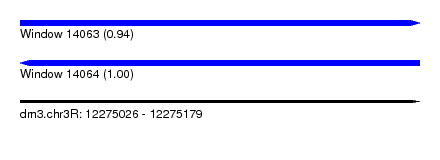
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,275,026 – 12,275,179 |
| Length | 153 |
| Max. P | 0.995235 |

| Location | 12,275,026 – 12,275,179 |
|---|---|
| Length | 153 |
| Sequences | 5 |
| Columns | 153 |
| Reading direction | forward |
| Mean pairwise identity | 76.00 |
| Shannon entropy | 0.43323 |
| G+C content | 0.39024 |
| Mean single sequence MFE | -39.87 |
| Consensus MFE | -14.41 |
| Energy contribution | -16.91 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.27 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.940458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
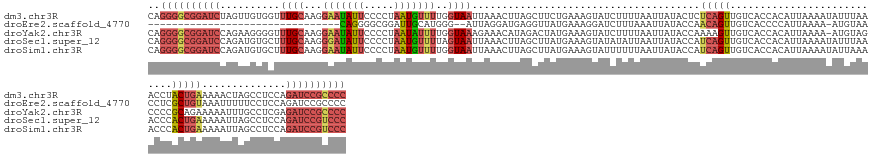
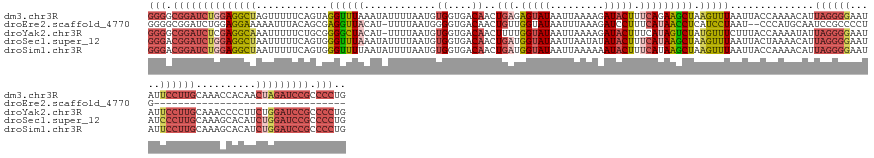
>dm3.chr3R 12275026 153 + 27905053 CAGGGGCGGAUCUAGUUGUGGUUUGCAAGGAAUAUUCCCCUAAUGUUUUGGUAAUUAAACUUAGCUUCUGAAAGUAUCUUUUAAUUAUACUCUCAGUUGUCACCACAUUAAAAUAUUUAAACCUACUGAAAAACUAGCCUCCAGAUCCGCCCC ..(((((((((((...((((((..(((((((....))).((((.((((........))))))))...((((.(((((.........))))).)))))))).))))))..................(((......))).....))))))))))) ( -41.50, z-score = -3.25, R) >droEre2.scaffold_4770 11806071 118 + 17746568 --------------------------------CAGGGGCGGAUUGCAUGGG--AUUAGGAUGAGGUUAUGAAAGGAUCUUUAAAUUAUACCAACAGUUGUCACCCCAUUAAAA-AUGUAACCUCGCUGUAAAUUUUUCCUCCAGAUCCGCCCC --------------------------------..((((((((((....(((--((((.(.(((((((((....((..............)).((....)).............-..))))))))).).)))....))))....)))))))))) ( -33.74, z-score = -1.57, R) >droYak2.chr3R 14692662 152 - 28832112 CAGGGGCGGAUCCAGAAGGGGUUUGCAAGGAAUAUUCCCCUAAUAUUUUGGUAAAGAAACAUAGACUAUGAAAGUAUCUUUUAAUUAUACCAAAAGUUGUCACCACAUUAAAA-AUGUAGCCCCGCAGAAAAAUUUGCCUCGAGAUCCGCCCC ..((((((((((..((.((((((.((((((........)))....((((((((.........(((.(((....))))))........))))))))..................-.)))))))))(((((....))))).))..)))))))))) ( -50.43, z-score = -4.98, R) >droSec1.super_12 251109 153 - 2123299 CAGGGGCGGAUCCAGAUGUGCUUUGCAAGGGAUAUUCCCCUAAUGUUUUAGUAAUUAAACUUAGCUUAUGAAAGUAUAUAUUAAUUAUACCAUCAGUUGUCACCACAUUAAAAUAUUUAAACCCACUGAAAAAUUAGCCUCCAGAUCCGUCCC ..((((((((((..((.(.(((.....((((......))))(((((((((((.........(((((.(((...(((((.......)))))))).))))).......)))))))))))..................))))))..)))))))))) ( -38.59, z-score = -3.12, R) >droSim1.chr3R 18329505 153 - 27517382 CAGGGGCGGAUCCAGAUGUGCUUUGCAAGGAAUAUUCCCCUAAUGUUUUGGUAAUUAAACUUAGCUUAUGAAAGUAUUUUUUAAUUAUACCAUCAGUUGUCACCACAUUAAAAUAUUAAAACCCACUGAAAAAUUAGCCUCCAGAUCCGUCCC ..((((((((((..((((((....(((((((....))).....((...(((((((((((....((((....))))....))))))..))))).)).))))...))))))................((((....))))......)))))))))) ( -35.10, z-score = -2.06, R) >consensus CAGGGGCGGAUCCAGAUGUGCUUUGCAAGGAAUAUUCCCCUAAUGUUUUGGUAAUUAAACUUAGCUUAUGAAAGUAUCUUUUAAUUAUACCAUCAGUUGUCACCACAUUAAAAUAUUUAAACCCACUGAAAAAUUAGCCUCCAGAUCCGCCCC ..((((((((((..........((((...(((((((.....)))))))..))))......................................(((((...........................)))))..............)))))))))) (-14.41 = -16.91 + 2.50)
| Location | 12,275,026 – 12,275,179 |
|---|---|
| Length | 153 |
| Sequences | 5 |
| Columns | 153 |
| Reading direction | reverse |
| Mean pairwise identity | 76.00 |
| Shannon entropy | 0.43323 |
| G+C content | 0.39024 |
| Mean single sequence MFE | -45.53 |
| Consensus MFE | -20.08 |
| Energy contribution | -23.20 |
| Covariance contribution | 3.12 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.995235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
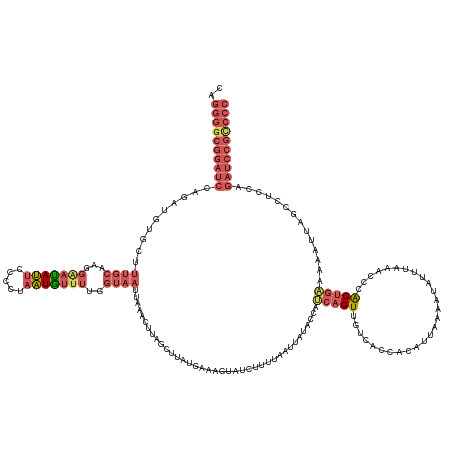
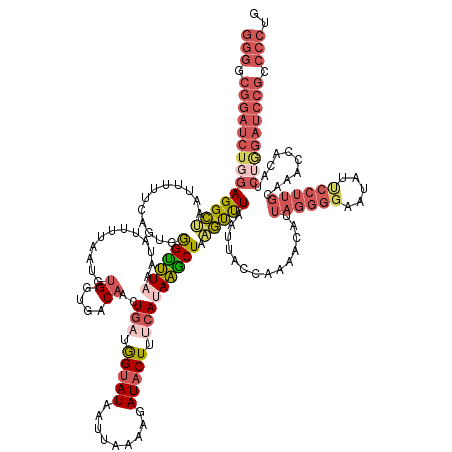
>dm3.chr3R 12275026 153 - 27905053 GGGGCGGAUCUGGAGGCUAGUUUUUCAGUAGGUUUAAAUAUUUUAAUGUGGUGACAACUGAGAGUAUAAUUAAAAGAUACUUUCAGAAGCUAAGUUUAAUUACCAAAACAUUAGGGGAAUAUUCCUUGCAAACCACAACUAGAUCCGCCCCUG (((((((((((((..(((........))).(((((........(((((((((((...((((((((((.........))))))))))((((...))))..)))))...))))))(((((....)))))..)))))....))))))))))))).. ( -54.60, z-score = -5.69, R) >droEre2.scaffold_4770 11806071 118 - 17746568 GGGGCGGAUCUGGAGGAAAAAUUUACAGCGAGGUUACAU-UUUUAAUGGGGUGACAACUGUUGGUAUAAUUUAAAGAUCCUUUCAUAACCUCAUCCUAAU--CCCAUGCAAUCCGCCCCUG-------------------------------- (((((((((.(((((((.(((((..(((((..(((((.(-(......)).)))))...)))))....)))))...((.....)).........))))...--.)))....)))))))))..-------------------------------- ( -32.50, z-score = -1.04, R) >droYak2.chr3R 14692662 152 + 28832112 GGGGCGGAUCUCGAGGCAAAUUUUUCUGCGGGGCUACAU-UUUUAAUGUGGUGACAACUUUUGGUAUAAUUAAAAGAUACUUUCAUAGUCUAUGUUUCUUUACCAAAAUAUUAGGGGAAUAUUCCUUGCAAACCCCUUCUGGAUCCGCCCCUG (((((((((((.((((((........)))((((((((((-.....)))))).......((((((((.......((((.((.............)).))))))))))))...(((((((...)))))))....))))))).))))))))))).. ( -50.63, z-score = -3.90, R) >droSec1.super_12 251109 153 + 2123299 GGGACGGAUCUGGAGGCUAAUUUUUCAGUGGGUUUAAAUAUUUUAAUGUGGUGACAACUGAUGGUAUAAUUAAUAUAUACUUUCAUAAGCUAAGUUUAAUUACUAAAACAUUAGGGGAAUAUCCCUUGCAAAGCACAUCUGGAUCCGCCCCUG (((.((((((..((.(((........((((((((((............((....))..(((.((((((.......)))))).))))))))).(((......)))....))))(((((.....)))))....)))...))..)))))).))).. ( -45.80, z-score = -3.35, R) >droSim1.chr3R 18329505 153 + 27517382 GGGACGGAUCUGGAGGCUAAUUUUUCAGUGGGUUUUAAUAUUUUAAUGUGGUGACAACUGAUGGUAUAAUUAAAAAAUACUUUCAUAAGCUAAGUUUAAUUACCAAAACAUUAGGGGAAUAUUCCUUGCAAAGCACAUCUGGAUCCGCCCCUG (((.((((((..((.(((......(((((..(((...((((....))))...))).)))))(((((..((((((.....(((....))).....)))))))))))......(((((((...)))))))...)))...))..)))))).))).. ( -44.10, z-score = -2.97, R) >consensus GGGGCGGAUCUGGAGGCUAAUUUUUCAGUGGGUUUAAAUAUUUUAAUGUGGUGACAACUGAUGGUAUAAUUAAAAGAUACUUUCAUAAGCUAAGUUUAAUUACCAAAACAUUAGGGGAAUAUUCCUUGCAAACCACAUCUGGAUCCGCCCCUG (((.((((((((((((((............((((((............((....))..(((.(((((.........))))).))))))))).)))))..............((((((.....))))))..........))))))))).))).. (-20.08 = -23.20 + 3.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:19:55 2011