| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,263,492 – 12,263,550 |
| Length | 58 |
| Max. P | 0.935782 |

| Location | 12,263,492 – 12,263,550 |
|---|---|
| Length | 58 |
| Sequences | 10 |
| Columns | 68 |
| Reading direction | forward |
| Mean pairwise identity | 66.30 |
| Shannon entropy | 0.70514 |
| G+C content | 0.54287 |
| Mean single sequence MFE | -16.66 |
| Consensus MFE | -10.68 |
| Energy contribution | -10.24 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.77 |
| Mean z-score | -0.51 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.935782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12263492 58 + 27905053 --------AACGAAAAUGUUUCAUUGU--UACCGCGGCCUGGGCAUUCUAGGGCUGCUCUCCCGCGAC --------...................--...(((((...(((((.(.....).)))))..))))).. ( -14.80, z-score = -0.08, R) >anoGam1.chr2R 35666864 68 - 62725911 GACGAAAGUCUUCCAGCGGUAUCUCCUCCGAACGGGGCCGUGGUAUGCGCGGGCUCGAUUCCCUGGAC (((....))).((((((((........)))..((((.(((((.....))))).)))).....))))). ( -29.60, z-score = -2.03, R) >droWil1.scaffold_181130 7913703 57 - 16660200 -----------GAAAAACUCAUAUUUUCAUACCUUGGCCUAGGCAUUCGAGGACUACUUUCGCGGGAU -----------(((((.......)))))...(((((((....))...)))))................ ( -8.40, z-score = 0.16, R) >droPer1.super_3 2261828 50 - 7375914 -----------------GGUUUAUUAUG-UACCGCGGCCUGGGCAUCCUUGGGCUGCUCUCACGCGAU -----------------(((........-.)))((((((..((....))..))))))........... ( -17.40, z-score = -1.40, R) >dp4.chr2 8460460 56 - 30794189 -----------AUUGGGGGUUUAUUAUG-UACCGCGGCCUGGGCAUCCUUGGGCUGCUCUCACGCGAU -----------..(((((((........-.)))((((((..((....))..)))))).))))...... ( -20.10, z-score = -1.22, R) >droAna3.scaffold_13340 15615995 57 + 23697760 -----------AAUAUAAAUGCUACCUAUUACCGCGGUCUGGGCAUCCUGGGGCUGUUGUCACGGGAU -----------.............((..(.((.(((((((.((....)).))))))).)).)..)).. ( -14.90, z-score = -0.08, R) >droEre2.scaffold_4770 11794526 63 + 17746568 ---AACGAAUAAAAGCUGUUUUAUUGU--UACCGCGGCCUGGGCAUUCUAGGGCUGCUCUCCCGCGAU ---(((.(((((((....)))))))))--)..(((((...(((((.(.....).)))))..))))).. ( -17.00, z-score = -0.30, R) >droYak2.chr3R 14681017 58 - 28832112 --------AACGAAAAUGUUUCAUUGU--UACCGCGGCCUGGGCAUUCUAGGGCUGCUCUCCCGCGAU --------...................--...(((((...(((((.(.....).)))))..))))).. ( -14.80, z-score = 0.04, R) >droSec1.super_12 239615 58 - 2123299 --------AACGAAAAUGUUUCAUUGU--UACCGCGGCCUGGGCAUUCUAGGGCUGCUCUCCCGCGAC --------...................--...(((((...(((((.(.....).)))))..))))).. ( -14.80, z-score = -0.08, R) >droSim1.chr3R 18317935 58 - 27517382 --------AACGAAAAUGUUUCAUUGU--UACCGCGGCCUGGGCAUUCUAGGGCUGCUCUCCCGCGAC --------...................--...(((((...(((((.(.....).)))))..))))).. ( -14.80, z-score = -0.08, R) >consensus ________A__GAAAAUGUUUCAUUGU__UACCGCGGCCUGGGCAUUCUAGGGCUGCUCUCCCGCGAU .......................((((......(((((((((.....))))).))))......)))). (-10.68 = -10.24 + -0.44)

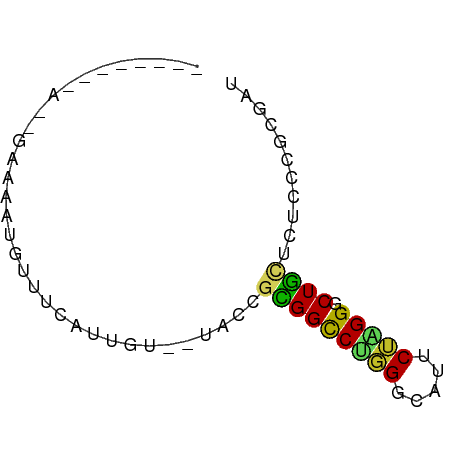
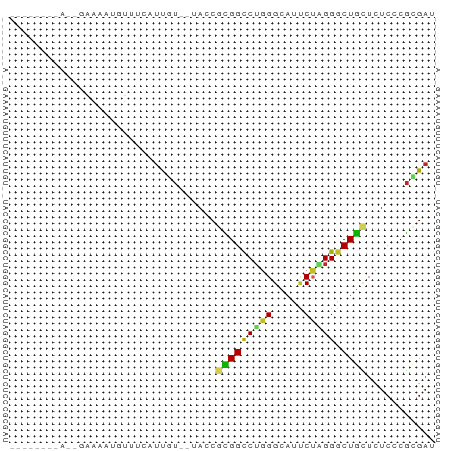
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:19:52 2011