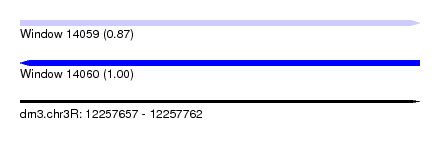
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,257,657 – 12,257,762 |
| Length | 105 |
| Max. P | 0.999360 |

| Location | 12,257,657 – 12,257,762 |
|---|---|
| Length | 105 |
| Sequences | 13 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 72.51 |
| Shannon entropy | 0.64547 |
| G+C content | 0.50322 |
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -20.25 |
| Energy contribution | -20.49 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.39 |
| Mean z-score | -0.50 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.865234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12257657 105 + 27905053 AUAUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGGGCCUAUUG----GAACACGA-UUUG-AGUGUGCCAGU--- ..((((.(((.((((((((..(((......)))...)))).......(((((((......)).))))))))).)))))))((----(.((((..-....-.)))).)))..--- ( -34.40, z-score = -1.02, R) >droSim1.chr3R 18312104 105 - 27517382 ACAUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGGGCCUAUUG----GAACACGA-UUUG-AGUGUGCCAGU--- ..((((.(((.((((((((..(((......)))...)))).......(((((((......)).))))))))).)))))))((----(.((((..-....-.)))).)))..--- ( -34.50, z-score = -0.82, R) >droSec1.super_12 233801 105 - 2123299 ACAUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGGGCCUAUUG----GAACACGA-UUUG-AGUGUGCCAGU--- ..((((.(((.((((((((..(((......)))...)))).......(((((((......)).))))))))).)))))))((----(.((((..-....-.)))).)))..--- ( -34.50, z-score = -0.82, R) >droYak2.chr3R 14674988 106 - 28832112 ACAUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGGGCCUACUG----GAACACAACUUUG-AGUGUCCCAUA--- (((.(((((..(((((.....)))))..))))))))...........(((((((......)).)))))...(((((..(((.----(((......))).-))))))))...--- ( -33.10, z-score = -0.48, R) >droEre2.scaffold_4770 11788495 105 + 17746568 ACAUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGGGCCUACUG----GAACACAA-UUAG-AGUGUUUCACU--- ...(((.(((.((((((((..(((......)))...)))).......(((((((......)).))))))))).)))))).((----((((((..-....-.))))))))..--- ( -34.20, z-score = -1.23, R) >droAna3.scaffold_13340 15608438 100 + 23697760 --AUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGGGCCUA-UG----GAGCA--A-UUGU-AACGCUCCAUU--- --((((.(((.((((((((..(((......)))...)))).......(((((((......)).))))))))).))))))-)(----((((.--.-....-...)))))...--- ( -35.20, z-score = -1.53, R) >dp4.chr2 8454154 101 - 30794189 ACAUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGGGCCUUUCU--ACAAGAAAUAUUGGUAUAU----------- .((.(((((..(((((.....)))))..)))))))(((.(((.(.(((...(((((((.......)))).)))..))).).)--)).))).............----------- ( -27.30, z-score = -0.13, R) >droPer1.super_3 6525622 98 + 7375914 AGUUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGGGCCUUCGU--CUAAGUGGAUAUAUUA-------------- ....(((((..(((((.....)))))..)))))((((((.(.....((((..((((((.......)))))...)..))))..--...).)))))).....-------------- ( -28.00, z-score = -0.05, R) >droWil1.scaffold_181130 3501423 88 + 16660200 --AUAGUGG-GAGGCAGUGUUGGUUAAGAU-UCAAUUAAGUCGGUGCUGGUUAAUGC-UCUGAGGCGCCGCGGAAUGUUCAA-UUAAAUUUCAA-------------------- --...((((-.((.(((..((((((((...-....)))).))))..))).))...((-(....))).))))(((((......-....)))))..-------------------- ( -18.70, z-score = 0.37, R) >droVir3.scaffold_13047 1974370 111 + 19223366 --AUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGGGCCUAUGG-GACGCGUACAAUUAAUUGUAGUUGUGUUUGC --((((.(((.((((((((..(((......)))...)))).......(((((((......)).))))))))).))))))).(-(((((((((((....)))))..))))))).. ( -34.50, z-score = -0.63, R) >droMoj3.scaffold_6540 3912064 94 + 34148556 --ACGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGGGCCUACACAGUUGAAUAUCCGC------------------ --..(((((..(((((.....)))))..)))))(((.(((.......(((((((......)).)))))((....))))).))).............------------------ ( -25.90, z-score = 0.73, R) >droGri2.scaffold_14906 8821625 111 + 14172833 --GUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCAUGAGUUCGAAUCUCGUCGGGGCCUAUUGUGGAAACCGCAAUUGAU-AAAAUUGUGUUCUU --..(((((..(((((.....)))))..)))))..((((...((.(((...(((((((.......))))).))..))).)).))))...(((((((...-..)))))))..... ( -29.40, z-score = -0.24, R) >anoGam1.chr2R 51288134 109 - 62725911 GUCUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGAGCCUUAGA---GUGUCACAAUUGGAUGAUCGCUCUGUA-- .((((((((..(((((.....)))))..))..((((.......))))(((((((......)).)))))))))))....((((---((((((........))).)))))))..-- ( -34.40, z-score = -0.71, R) >consensus A_AUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGGGCCUAUUG____GAACACAA_UUUG_AGUGU_CCA_U___ ....((.(((.((((((((..(((......)))...)))).......(((((((......)).))))))))).))))).................................... (-20.25 = -20.49 + 0.25)

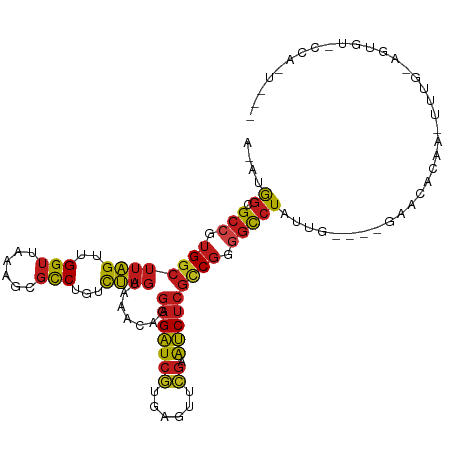
| Location | 12,257,657 – 12,257,762 |
|---|---|
| Length | 105 |
| Sequences | 13 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 72.51 |
| Shannon entropy | 0.64547 |
| G+C content | 0.50322 |
| Mean single sequence MFE | -28.39 |
| Consensus MFE | -19.27 |
| Energy contribution | -20.10 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.82 |
| SVM RNA-class probability | 0.999360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12257657 105 - 27905053 ---ACUGGCACACU-CAAA-UCGUGUUC----CAAUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAUAU ---..(((.((((.-....-..)))).)----))...(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).... ( -31.90, z-score = -3.11, R) >droSim1.chr3R 18312104 105 + 27517382 ---ACUGGCACACU-CAAA-UCGUGUUC----CAAUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAUGU ---..(((.((((.-....-..)))).)----))...(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).... ( -31.90, z-score = -2.57, R) >droSec1.super_12 233801 105 + 2123299 ---ACUGGCACACU-CAAA-UCGUGUUC----CAAUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAUGU ---..(((.((((.-....-..)))).)----))...(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).... ( -31.90, z-score = -2.57, R) >droYak2.chr3R 14674988 106 + 28832112 ---UAUGGGACACU-CAAAGUUGUGUUC----CAGUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAUGU ---..((((((((.-.......))))))----))...(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).... ( -36.90, z-score = -2.96, R) >droEre2.scaffold_4770 11788495 105 - 17746568 ---AGUGAAACACU-CUAA-UUGUGUUC----CAGUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAUGU ---(.((.(((((.-....-..))))).----)).).(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).... ( -30.30, z-score = -1.83, R) >droAna3.scaffold_13340 15608438 100 - 23697760 ---AAUGGAGCGUU-ACAA-U--UGCUC----CA-UAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAU-- ---.((((((((..-....-.--)))))----))-).(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))..-- ( -36.70, z-score = -4.34, R) >dp4.chr2 8454154 101 + 30794189 -----------AUAUACCAAUAUUUCUUGU--AGAAAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAUGU -----------...........((((....--.))))(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).... ( -26.10, z-score = -1.97, R) >droPer1.super_3 6525622 98 - 7375914 --------------UAAUAUAUCCACUUAG--ACGAAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAACU --------------................--.....(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).... ( -25.20, z-score = -2.40, R) >droWil1.scaffold_181130 3501423 88 - 16660200 --------------------UUGAAAUUUAA-UUGAACAUUCCGCGGCGCCUCAGA-GCAUUAACCAGCACCGACUUAAUUGA-AUCUUAACCAACACUGCCUC-CCACUAU-- --------------------((((.((((((-((((........(((.((......-..........)).)))..))))))))-)).)))).............-.......-- ( -9.39, z-score = -0.16, R) >droVir3.scaffold_13047 1974370 111 - 19223366 GCAAACACAACUACAAUUAAUUGUACGCGUC-CCAUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAU-- ...........(((((....)))))......-.....(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))..-- ( -26.50, z-score = -1.85, R) >droMoj3.scaffold_6540 3912064 94 - 34148556 ------------------GCGGAUAUUCAACUGUGUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCGU-- ------------------((((........))))...(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))..-- ( -26.90, z-score = -0.89, R) >droGri2.scaffold_14906 8821625 111 - 14172833 AAGAACACAAUUUU-AUCAAUUGCGGUUUCCACAAUAGGCCCCGACGAGAUUCGAACUCAUGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAC-- ..............-....((((.((...)).)))).(((.(((..((((((.........))))))(((((.....)))))(.((((.........))))).))).)))..-- ( -23.30, z-score = -1.39, R) >anoGam1.chr2R 51288134 109 + 62725911 --UACAGAGCGAUCAUCCAAUUGUGACAC---UCUAAGGCUCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAGAC --...((((.(.((((......))))).)---)))..(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).... ( -32.10, z-score = -2.70, R) >consensus ___A_UGG_ACACU_AAAA_UUGUGUUC____CAAUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAU_U .....................................(((.(((((((((.(((......)))))))(((((.....)))))))((((.........))))..))).))).... (-19.27 = -20.10 + 0.83)
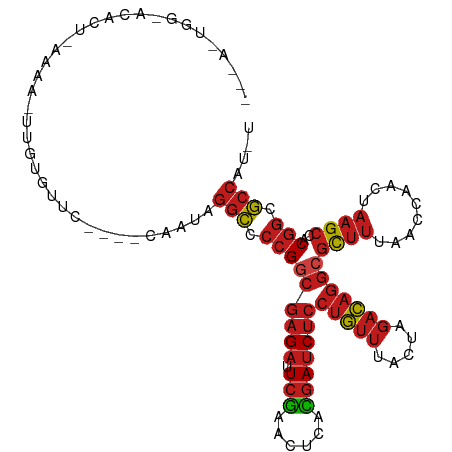
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:19:51 2011