| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,254,023 – 12,254,129 |
| Length | 106 |
| Max. P | 0.780991 |

| Location | 12,254,023 – 12,254,129 |
|---|---|
| Length | 106 |
| Sequences | 13 |
| Columns | 124 |
| Reading direction | forward |
| Mean pairwise identity | 70.65 |
| Shannon entropy | 0.62370 |
| G+C content | 0.39765 |
| Mean single sequence MFE | -22.47 |
| Consensus MFE | -9.90 |
| Energy contribution | -11.18 |
| Covariance contribution | 1.29 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.780991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
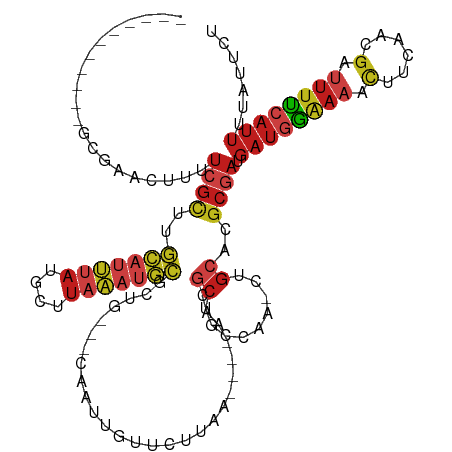
>dm3.chr3R 12254023 106 + 27905053 -----------GCGGAUUUUUCGCUAGCAUUUAUGCUUAAAUGCGCUG---CAAUUGUUCUUAA----AACGCUGCCAACCUGCACGCGAUGAUGGAAAACCUCAACGAUUUCCAUUUUAUUCU -----------..((((..((.((..(((((((....)))))))...)---)))..))))....----..((((((......))).)))..((((((((.(......).))))))))....... ( -25.00, z-score = -1.64, R) >droSim1.chr3R 18308376 106 - 27517382 -----------GCGGAUUUUUCGCUAGCAUUUAUGCUUAAAUGCGCUG---CAAUUGUUCUUAA----AACGCUGCCAACCUGCACGCGAUGAUGGAAAACCUCAACGAUUUCCAUUUUAUUCU -----------..((((..((.((..(((((((....)))))))...)---)))..))))....----..((((((......))).)))..((((((((.(......).))))))))....... ( -25.00, z-score = -1.64, R) >droSec1.super_12 230086 106 - 2123299 -----------GCGGAUUUUUCGCUAGCAUUUAUGCUUAAAUGCGCUG---CAAUUGUUCUUAA----AACGCUGCCAACCUGCACGCGAUGAUGGAAAACCUCAACGAUUUCCAUUUUAUUCU -----------..((((..((.((..(((((((....)))))))...)---)))..))))....----..((((((......))).)))..((((((((.(......).))))))))....... ( -25.00, z-score = -1.64, R) >droYak2.chr3R 14671202 106 - 28832112 -----------GCGGAGUUUUCGCUAGCAUUUAUGCUUAAAUGCGAUG---CAAUUGUUCUUAA----AACGCUGCCAACCUGCACGCGAUGAUGGAAAACCUCAACGAUUUUCAUUUUAUUCU -----------..(..((((((((..(((((((....)))))))...)---)............----..((((((......))).)))......))))))..).................... ( -24.60, z-score = -1.21, R) >droEre2.scaffold_4770 11784800 106 + 17746568 -----------GCGCAGUUUUCGCUAGCAUUUAUGCUUAAAUGCGCUG---CAAUUGUUCUUAA----AACGCUGCCAACCUGCACGCGAUGAUGGAAAACCUCAACGAUUUUCAUUUUAUUCU -----------..(((((........(((((((....)))))))))))---)((((((......----..((((((......))).))).(((.((....)))))))))))............. ( -23.00, z-score = -0.51, R) >droAna3.scaffold_13340 15605245 106 + 23697760 -----------GCAAACUUUUCGCUUGCAUUUAUGCUUAAAUGCACUG---CAAUUGUUCUUAA----AACGCUGCCAGGCUGCACGCGAUGAUGGAAAACUUCAACGAUUUUCAUUUUAUUCU -----------.........((((.((((((((....)))))))).((---((...(((.....----)))(((....))))))).)))).((((((((.(......).))))))))....... ( -21.80, z-score = -0.69, R) >dp4.chr2 8450224 98 - 30794189 -----------GCGAACUUUUCGCCUGCAUUUAUGCUUAAAUGCCCAA---CAAUUGUUCUUAA----AACGC--------UGCACGCGAUGAUGGAAAACUUCAACGAUUUUCAUUUUAUACU -----------..((((..((.....(((((((....)))))))....---.))..))))....----..(((--------.....)))..((((((((.(......).))))))))....... ( -16.80, z-score = -1.18, R) >droPer1.super_3 2251174 98 - 7375914 -----------GCGAACUUUUCGCCUGCAUUUAUGCUUAAAUGCCCAA---CAAUUGUUCUUAA----AACGC--------UGCACGCGAUGAUGGAAAACUUCAACGAUUUUCAUUUUAUACU -----------..((((..((.....(((((((....)))))))....---.))..))))....----..(((--------.....)))..((((((((.(......).))))))))....... ( -16.80, z-score = -1.18, R) >droWil1.scaffold_181108 2396943 101 + 4707319 -----------ACUAAUUUUCAGUAAACAUAAAGUUUUGUUACCAAUCUGAAGAGGGAGGGGAAUUGCAAUCGGGUGAAGGUAUAUGCAAAAGUCAGUUGCCA-AAUAAUUUU----------- -----------.....(((((((..((((........))))......)))))))((.((..((.((((((((.......)))...)))))...))..)).)).-.........----------- ( -15.80, z-score = 0.64, R) >droGri2.scaffold_14906 8816895 97 + 14172833 -----------CUGAACUUUUCGCCUGCAUUUAUGCUUAAAUGCACUG---CAAUUGUUUCUAA----AAUGCAGCCA--CUGCACGCGAUGAUGGAAAACUUCAACGAUUUUCAUU------- -----------.........((((.((((....(((......)))(((---((.((.......)----).)))))...--.)))).)))).((((((((.(......).))))))))------- ( -22.00, z-score = -1.94, R) >droMoj3.scaffold_6540 3906159 101 + 34148556 -------------GAACUUUUCGCCUGCAUUUAUGCUUAAAUGCGCUG---CAAUUGUUCCUAA----AAUGCAGCCG--CGGCACGCGAUGAUGGAAAACUUGAACGAUUUUCAUUUAUUUU- -------------.......((((.((((((((....)))))((((((---((.((.......)----).))))).))--).))).)))).((((((((.(......).))))))))......- ( -25.50, z-score = -1.48, R) >droVir3.scaffold_13047 1969682 97 + 19223366 -------------GAACUUUUCGCCUGCAUUUAUGCUUAAAUGCGCUG---CAAUUGUUCCUAA----AAUGCAGCCA--CUGCACGCGAUGAUGGAAAACUUGAACGAUUUUCAUUUC----- -------------.......((((.((((.....((......))((((---((.((.......)----).))))))..--.)))).)))).((((((((.(......).))))))))..----- ( -25.00, z-score = -2.61, R) >anoGam1.chr3R 19479252 121 - 53272125 UUUUGUUUCCUUCCAACACUUUAAUUAAAUCCUUCGUUACUCUUGUUUGGUCACCUGUUUCGAGCGGUGACACUACAUCGGUGAACGGCAGGAUAGCACAUUU--AUGUUUUCCAUUUUUCCC- ............................(((((((((((((..(((...((((((.((.....))))))))...)))..)))).)))).))))).........--..................- ( -25.80, z-score = -1.55, R) >consensus ___________GCGAACUUUUCGCUUGCAUUUAUGCUUAAAUGCGCUG___CAAUUGUUCUUAA____AACGCUGCCAA_CUGCACGCGAUGAUGGAAAACUUCAACGAUUUUCAUUUUAUUCU ....................((((..(((((((....)))))))...........................((.........))..)))).((((((((.(......).))))))))....... ( -9.90 = -11.18 + 1.29)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:19:49 2011