
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,246,248 – 12,246,346 |
| Length | 98 |
| Max. P | 0.887078 |

| Location | 12,246,248 – 12,246,346 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 66.30 |
| Shannon entropy | 0.64277 |
| G+C content | 0.52961 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -14.32 |
| Energy contribution | -14.01 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.887078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
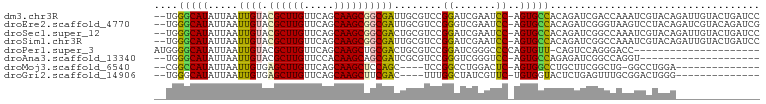
>dm3.chr3R 12246248 98 + 27905053 --UGGGCAUAUUAAUUGUACGCUUGUUCAGCAAGCGGCGAUUGCGUCCGGAUCGAAUCC-AGUGCCACAGAUCGACCAAAUCGUACAGAUUGUACUGAUCC --(((((((..(((((((.((((((.....))))))))))))).....((((...))))-.))))).))(((((....((((.....))))....))))). ( -30.90, z-score = -1.98, R) >droEre2.scaffold_4770 11774948 98 + 17746568 --UGGGCAUAUUAAUUGUACGCUUGUUCAGCAAGCGGCGAUUGCGUCCGGGUCGAAUCC-AGUGCCACAGAUCGGGUAAGUCCUACAGAUCGUACAGAUCG --(((((((..(((((((.((((((.....))))))))))))).....((((...))))-.))))).))((((..(((.(((.....)))..))).)))). ( -30.60, z-score = -1.26, R) >droSec1.super_12 222286 98 - 2123299 --UGGGCAUAUUAAUUGUACGCUUGUUCAGCAAGCGGCGACUGCGUCCGGAUCGAAUCC-AGUGCCACAGAUCGGCCAAAUCGUACAGAUUGUACUGAUCC --(((((((.....((((.((((((.....))))))))))........((((...))))-.))))).))((((((...((((.....))))...)))))). ( -31.10, z-score = -1.49, R) >droSim1.chr3R 18300593 98 - 27517382 --UGGGCAUAUUAAUUGUACGCUUGUUCAGCAAGCGGCGAUUGCGUCCGGAUCGAAUCC-AGUGCCACAGAUCGGCCAAAUCGUACAGAUUGUACUGAUCC --(((((((..(((((((.((((((.....))))))))))))).....((((...))))-.))))).))((((((...((((.....))))...)))))). ( -34.00, z-score = -2.49, R) >droPer1.super_3 2242015 79 - 7375914 AUGGGGCAUAUUAAUUGUACGCUUGUUCAGCAAGCUGCGACUGCGUCCGGAUCGGGCCCCAGUGUU-CAGUCCAGGGACC--------------------- .((((((.......(((((.(((((.....))))))))))......(((...))))))))).....-..(((....))).--------------------- ( -27.30, z-score = -0.80, R) >droAna3.scaffold_13340 15598088 78 + 23697760 --UGGGCAUAUUAAUUGUACGCUUGUUCCACAAGCAGCGAUCGCGUCCGGGUCGGGUCC-AGUGCCAGAGAUCGGCCAGGU-------------------- --.((((.............(((((.....))))).((....)))))).((((((.((.-.......))..))))))....-------------------- ( -23.30, z-score = 0.16, R) >droMoj3.scaffold_6540 3896725 79 + 34148556 --CGGCCAUAUUAAUUGUGAGCUUGUUCAGCAAGCUCCAGC----UCCGGCCUGGACUC-AGUGGCCUGCUUCGGCUG-GGCCUGGA-------------- --.((((.......(((.(((((((.....)))))))))).----...)))).....((-...(((((((....)).)-))))..))-------------- ( -32.30, z-score = -1.06, R) >droGri2.scaffold_14906 8805753 80 + 14172833 --UGGGCAUAUUAAUUGUGAGCUUGUUCAGCAAGCUUCGAC----UUUGGCUAUCGUUC-UGUGGUACUCUGAGUUUGCGGACUGGG-------------- --.((((.(((.....))).))))((((.((((((((.((.----....(((((.....-.)))))..)).))))))))))))....-------------- ( -19.50, z-score = 0.08, R) >consensus __UGGGCAUAUUAAUUGUACGCUUGUUCAGCAAGCGGCGACUGCGUCCGGAUCGAAUCC_AGUGCCACAGAUCGGCCAAGUCCUACA______________ ....(((((..........((((((.....)))))).((((((....))).))).......)))))................................... (-14.32 = -14.01 + -0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:19:48 2011