| Sequence ID | dm3.chr3R |
|---|---|
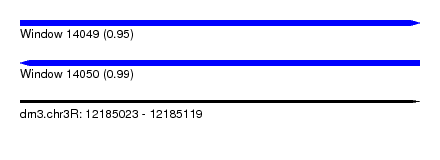
| Location | 12,185,023 – 12,185,119 |
| Length | 96 |
| Max. P | 0.994290 |

| Location | 12,185,023 – 12,185,119 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 73.85 |
| Shannon entropy | 0.50508 |
| G+C content | 0.46851 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -18.56 |
| Energy contribution | -19.57 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.945566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
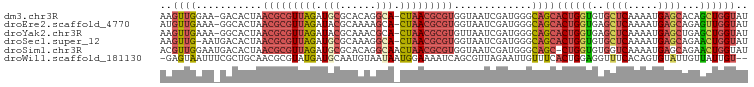
>dm3.chr3R 12185023 96 + 27905053 AAGUUGGAA-GACACUAACGCGUUAGAUGCGCACAGGCA-CUAACGCGUGGUAAUCGAUGGGCAGCACUGGUGUGCUCAAAAUGAGCACAGCUGGUAU ..(((.(..-((.((((.((((((((.(((......)))-))))))))))))..))..).)))...((..((((((((.....))))))).)..)).. ( -37.20, z-score = -3.22, R) >droEre2.scaffold_4770 11712324 96 + 17746568 AUGUUGAAA-GGCACUAACGCGUUAGAUACGCAAAAGCA-CUAACGCGUGGUAAUCGAUGGGCAGCACUGGUGAGCUCAAAAUGAGCAGAGUUGGUAU ..(((((..-.((....(((((((((....((....)).-))))))))).))..))))).......(((..(..((((.....))))...)..))).. ( -26.10, z-score = -0.73, R) >droYak2.chr3R 14600396 96 - 28832112 AAGUUGAAA-GGCACUAACGCGUUAGAUACGCAAACGCA-CUAACGCGUGUUAAUCGAUGGGCAGCACUGGUGAGCUCAAAAUGAGCUGAGCUGGUAU ..((((...-((.....(((((((((....((....)).-))))))))).....))......))))((..((.(((((.....)))))..))..)).. ( -30.30, z-score = -1.65, R) >droSec1.super_12 160948 96 - 2123299 AAGUUG-AAUGACACUAACGCGUUAGAUGCGCAAAGGCA-CUAACGCGUGGUAAUCGAUGGGCAGCACUGGUGUGCUCAAAAUGAGCAGAACUGGUAU ..((((-.((((.((((.((((((((.(((......)))-))))))))))))..)).))...))))((..((.(((((.....)))))..))..)).. ( -35.40, z-score = -3.51, R) >droSim1.chr3R 18239024 97 - 27517382 ACGUUGGAAUGACACUAACGCGUUAGAUGCGCACAGGCAACUAACGCGUGGUAAUCGAUGGGCAGC-CUGGUGUGGUCAAAAUGAGCAGAACUGGUAU .((((((....((....(((((((((.(((......))).))))))))).))..))))))....((-(.(((.((.((.....)).))..)))))).. ( -31.00, z-score = -1.40, R) >droWil1.scaffold_181130 3412160 95 + 16660200 -GAGUAAUUUCGCUGCAACGCGUAUGAUGCAAUGUAAUAAUGGAAAAUCAGCGUUAGAAUUGUUUCACUGGAGGUUUCACAGUGUAUUGUUAUUGU-- -.((((((..(((((....((.(.(((.(((((....(((((.........)))))..))))).)))....).))....)))))....))))))..-- ( -16.10, z-score = 1.46, R) >consensus AAGUUGAAA_GACACUAACGCGUUAGAUGCGCAAAGGCA_CUAACGCGUGGUAAUCGAUGGGCAGCACUGGUGUGCUCAAAAUGAGCAGAGCUGGUAU ..((((...........(((((((((.(((......))).))))))))).............))))((((((..((((.....))))...)))))).. (-18.56 = -19.57 + 1.01)
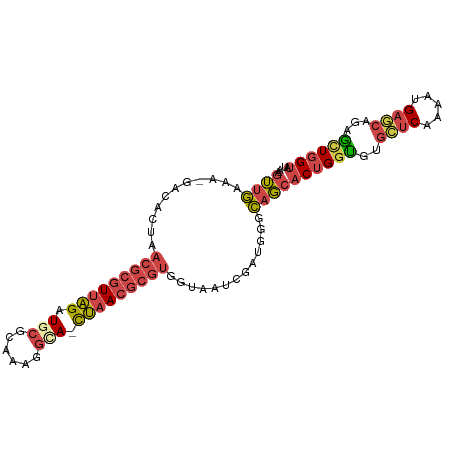
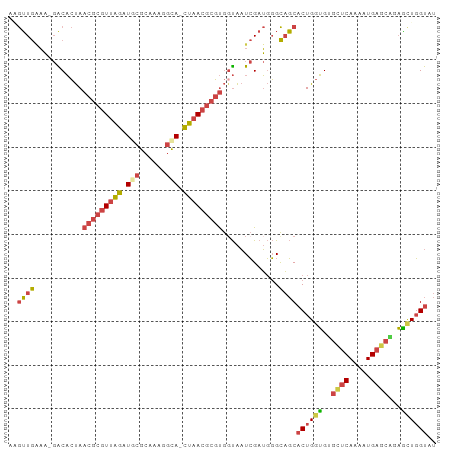
| Location | 12,185,023 – 12,185,119 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 73.85 |
| Shannon entropy | 0.50508 |
| G+C content | 0.46851 |
| Mean single sequence MFE | -24.69 |
| Consensus MFE | -14.74 |
| Energy contribution | -16.05 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.994290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12185023 96 - 27905053 AUACCAGCUGUGCUCAUUUUGAGCACACCAGUGCUGCCCAUCGAUUACCACGCGUUAG-UGCCUGUGCGCAUCUAACGCGUUAGUGUC-UUCCAACUU ....(((((((((((.....))))))).....))))......(((....(((((((((-(((......))).)))))))))....)))-......... ( -32.90, z-score = -3.75, R) >droEre2.scaffold_4770 11712324 96 - 17746568 AUACCAACUCUGCUCAUUUUGAGCUCACCAGUGCUGCCCAUCGAUUACCACGCGUUAG-UGCUUUUGCGUAUCUAACGCGUUAGUGCC-UUUCAACAU ...........((((.....))))......(..(((.............(((((((((-(((......))).))))))))))))..).-......... ( -22.91, z-score = -2.03, R) >droYak2.chr3R 14600396 96 + 28832112 AUACCAGCUCAGCUCAUUUUGAGCUCACCAGUGCUGCCCAUCGAUUAACACGCGUUAG-UGCGUUUGCGUAUCUAACGCGUUAGUGCC-UUUCAACUU ....((((.((((((.....))))).....).))))......((...(((((((((((-((((....)))).)))))))))..))...-..))..... ( -28.50, z-score = -2.92, R) >droSec1.super_12 160948 96 + 2123299 AUACCAGUUCUGCUCAUUUUGAGCACACCAGUGCUGCCCAUCGAUUACCACGCGUUAG-UGCCUUUGCGCAUCUAACGCGUUAGUGUCAUU-CAACUU ....((((.((((((.....)))).....)).))))......(((....(((((((((-(((......))).)))))))))....)))...-...... ( -27.10, z-score = -3.00, R) >droSim1.chr3R 18239024 97 + 27517382 AUACCAGUUCUGCUCAUUUUGACCACACCAG-GCUGCCCAUCGAUUACCACGCGUUAGUUGCCUGUGCGCAUCUAACGCGUUAGUGUCAUUCCAACGU ....((((.(((................)))-))))......(((....(((((((((.(((......))).)))))))))....))).......... ( -22.69, z-score = -1.03, R) >droWil1.scaffold_181130 3412160 95 - 16660200 --ACAAUAACAAUACACUGUGAAACCUCCAGUGAAACAAUUCUAACGCUGAUUUUCCAUUAUUACAUUGCAUCAUACGCGUUGCAGCGAAAUUACUC- --............(((((.((....)))))))............(((((..............((.(((.......))).))))))).........- ( -14.03, z-score = -1.00, R) >consensus AUACCAGCUCUGCUCAUUUUGAGCACACCAGUGCUGCCCAUCGAUUACCACGCGUUAG_UGCCUUUGCGCAUCUAACGCGUUAGUGCC_UUUCAACUU ....((((..(((((.....))))).......)))).............(((((((((.(((......))).)))))))))................. (-14.74 = -16.05 + 1.31)
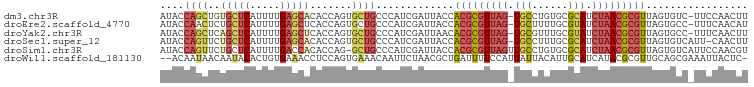

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:19:42 2011