| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,156,059 – 12,156,166 |
| Length | 107 |
| Max. P | 0.656814 |

| Location | 12,156,059 – 12,156,166 |
|---|---|
| Length | 107 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.32 |
| Shannon entropy | 0.70392 |
| G+C content | 0.45331 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -8.14 |
| Energy contribution | -7.95 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.656814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
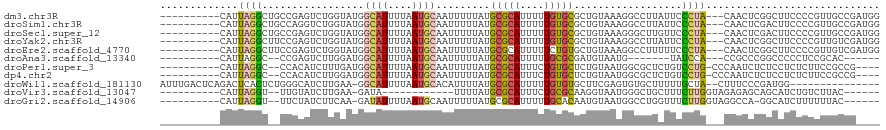
>dm3.chr3R 12156059 107 - 27905053 ----------CAUUAGGCUGCCGAGUCUGGUAUGGCAUUUUAAUGCAAUUUUUAUGCGCAUUUUUGUGCGCUGUAAAGGCCUUAUUCCCUA---CAACUCGGCUUCCCCGUUGCCGAUGG ----------((((.(((.(((((((...(((.(((((....))))..(((((((((((((....)))))).)))))))........).))---).))))))).........))))))). ( -33.30, z-score = -1.86, R) >droSim1.chr3R 18208901 107 + 27517382 ----------CAUUAGGCUGCCAGGUCUGGUAUGGCAUUUUAAUGCAAUUUUUAUGCGCAUUUUUGUGCGCUGUAAAGGCCUUAUUCCCUA---CAACUCGACUUCCCCGUUGCCGAUGG ----------((((.(((.((.(((((.(((.((((((....))))..(((((((((((((....)))))).)))))))............---))))).)))))....)).))))))). ( -28.80, z-score = -0.81, R) >droSec1.super_12 132395 107 + 2123299 ----------CAUUAGGCUGCCGAGUCUGGUAUGGCAUUUUAAUGCAAUUUUUAUGCGCAUUUUUGUGCGCUGUAAAGGGCUUGUUCCCUA---CAACUCGACUUCCCCGUUGCCGAUGG ----------((((.(((.((.(((((.(((.((((((....)))).....((((((((((....)))))).))))((((......)))).---))))).)))))....)).))))))). ( -31.60, z-score = -1.38, R) >droYak2.chr3R 14571476 107 + 28832112 ----------CAUUAGGCUUCCGAGUCUGGUAUGGCAUUUUAAUGCAAUUUUUAUGCGCAUUUUUGUGCGCUGUAAAGGCCUUAUUCCCUA---CAACUCGGCUUCCCCGUUGUCGAUGG ----------.....((...((((((...(((.(((((....))))..(((((((((((((....)))))).)))))))........).))---).))))))...))(((((...))))) ( -27.70, z-score = -0.89, R) >droEre2.scaffold_4770 11681771 107 - 17746568 ----------CAUUAGGCUUCCGAGUCUGGUAUGGCAUUUUAAUGCAAUUUUUAUGCGCAUUUUUCUGUGCUGUAAAGGCCUUUUUCCCUA---CAACUCGGCUUCCCCGUUGUCGAUGG ----------.....((...((((((.((.((((((((...(((((...........))))).....)))))))).(((........))).---))))))))...))(((((...))))) ( -24.60, z-score = -0.46, R) >droAna3.scaffold_13340 15507920 92 - 23697760 ----------CAUUAGGC--CCGAGUCUUGGAUGGCAUUUUAAUGCAAUUUUUAUGCGCAUUUUUGCGCGAUGUAAUG-------UAUCCA---CCGCCCGGCCCCCUCCGCAC------ ----------.....(((--(.(.((..((((((((((....))))........((((((....))))))........-------))))))---..))).))))..........------ ( -27.70, z-score = -1.74, R) >droPer1.super_3 2139415 103 + 7375914 ----------CAUUAGGC--CCACAUCUUUGAUGGCAUUUUAAUGCAAUUUUUAUGCGCAUUUCUGUGCUCUGUAAUGGCGCUCUGUCCUG-CCCAAUCUCUCCUCUCUUCCGCCG---- ----------.....(((--...((((...))))((((...(((((...........)))))...)))).......(((.((........)-))))................))).---- ( -20.20, z-score = -0.99, R) >dp4.chr2 8339049 103 + 30794189 ----------CAUUAGGC--CCACAUCUUGGAUGGCAUUUUAAUGCAAUUUUUAUGCGCAUUUCUGUGCUCUGUAAUGGCGCUCUGUCCUG-CCCAAUCUCUCCUCUCUUCCGCCG---- ----------.....(((--.........(((.(((((...(((((...........)))))...)))))......(((.((........)-)))).....)))........))).---- ( -21.53, z-score = -0.76, R) >droWil1.scaffold_181130 3363711 102 - 16660200 AUUUGACUCAGACUCACUCUGGGCAUCUUGAA-GGCAUUUUAAUGCACAUUUUAUGCGCAUUUUUGUGUGCUUCGAGUGUGCUUUUUGCUA--CUUUCCCGAUGG--------------- ......((((((.....))))))((((..(((-((.........((((((((...((((((....))))))...)))))))).........--)))))..)))).--------------- ( -32.07, z-score = -3.70, R) >droVir3.scaffold_13047 1782106 89 - 19223366 ----------CAUUAGGU--UUGUAUCUUGAA-GAUA------------UUUUAUGCGCAUUUCUGCGCAAGGUAAUGGGCUGCUUUCUUGGUAGAGAGCAGCAUCUGUCUUAC------ ----------(((((...--..(((((.....-))))------------)....((((((....))))))...))))).((((((((((....))))))))))...........------ ( -29.80, z-score = -3.05, R) >droGri2.scaffold_14906 12601355 100 - 14172833 ----------CAUUAGGU--UUCUAUCUUCAA-GAUAUUUUAAUGCAAUUUUUAUGCGCAUUUUUGCACAAUGUAAUGGCCUGGUUUCUUGGUAGGCCA-GGCAUCUUUUUUAC------ ----------(((((.((--(..((((.....-)))).......(((.......)))(((....)))..))).)))))(((((((((......))))))-)))...........------ ( -24.10, z-score = -2.21, R) >consensus __________CAUUAGGC__CCGAGUCUUGUAUGGCAUUUUAAUGCAAUUUUUAUGCGCAUUUUUGUGCGCUGUAAAGGCCUUAUUCCCUA___CAACUCGGCUUCCCCGUUGCCG____ .............((((.................((((....)))).........(((((....)))))..................))))............................. ( -8.14 = -7.95 + -0.18)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:19:37 2011