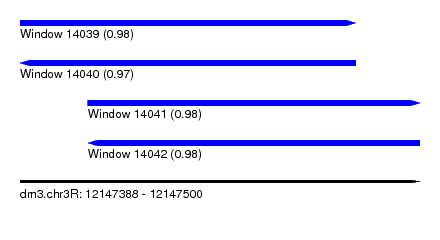
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,147,388 – 12,147,500 |
| Length | 112 |
| Max. P | 0.983683 |

| Location | 12,147,388 – 12,147,482 |
|---|---|
| Length | 94 |
| Sequences | 13 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.88 |
| Shannon entropy | 0.46120 |
| G+C content | 0.56976 |
| Mean single sequence MFE | -33.87 |
| Consensus MFE | -27.51 |
| Energy contribution | -27.46 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
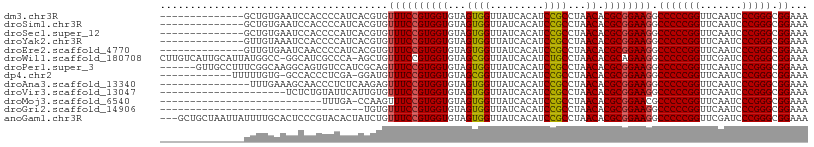
>dm3.chr3R 12147388 94 + 27905053 --------------GCUGUGAAUCCACCCCAUCACGUGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAA --------------..(((((..((((..(((((((......)))))))..))))...)))))(((((.......)))))...(((((((.......))))).))... ( -40.20, z-score = -3.42, R) >droSim1.chr3R 18200150 94 - 27517382 --------------GCUGUGAAUCCACCCCAUCACGUGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAA --------------..(((((..((((..(((((((......)))))))..))))...)))))(((((.......)))))...(((((((.......))))).))... ( -40.20, z-score = -3.42, R) >droSec1.super_12 123702 94 - 2123299 --------------GCUGUGAAUCCACCCCAUCACGUGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAA --------------..(((((..((((..(((((((......)))))))..))))...)))))(((((.......)))))...(((((((.......))))).))... ( -40.20, z-score = -3.42, R) >droYak2.chr3R 14562336 94 - 28832112 --------------GUUGUAAAUCCACCCCAUCACGUGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAA --------------((.((((..((((..(((((((......)))))))..)))))))).)).(((((.......)))))...(((((((.......))))).))... ( -36.00, z-score = -2.66, R) >droEre2.scaffold_4770 11672693 94 + 17746568 --------------GUUGUGAAUCAACCCCAUCACGUGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAA --------------..(((((((((.(..(((((((......)))))))..)))))..)))))(((((.......)))))...(((((((.......))))).))... ( -34.60, z-score = -1.88, R) >droWil1.scaffold_180708 3501503 106 + 12563649 CUUGUCAUUGCAUUAUGGCC-GGCAUCGCCCA-AGCUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAA ....((.(((((((((((.(-(((........-.))))...))))))))))).))......(.(((((.......))))).).(((((((.......))))).))... ( -32.40, z-score = 0.65, R) >droPer1.super_3 6521062 102 + 7375914 ------GUUGCCUUUCGGCAAGGCAGUGUCCAUCGCAGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAA ------.(((((....)))))(((.(((.....)))..((((((((((...((((.........))))...)).)))))))))))(((((.......)))))...... ( -37.00, z-score = -1.18, R) >dp4.chr2 8330741 94 - 30794189 ------------UUUUUGUG-GCCACCCUCGA-GGAUGUUUCCGUGGUGUAGCGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAA ------------.(((((.(-(((..((....-))....(((((((((...((((.........))))...)).)))))))))))(((((.......)))))))))). ( -35.50, z-score = -0.99, R) >droAna3.scaffold_13340 15499175 93 + 23697760 ---------------UUUGAAAGCAACCCUCUCAAGAGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAA ---------------(((((.((.....)).)))))..((((((((((...((((.........))))...)).)))))))).(((((((.......))))).))... ( -29.70, z-score = -1.00, R) >droVir3.scaffold_13047 1772924 87 + 19223366 ---------------------UCUCUGUAUUCAUUGUGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAA ---------------------.................((((((((((...((((.........))))...)).)))))))).(((((((.......))))).))... ( -27.90, z-score = -1.01, R) >droMoj3.scaffold_6540 18740883 80 - 34148556 ---------------------------UUUGA-CCAAGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAACGCCCCCGGUUCAAUCCCGGGCGGAAA ---------------------------.....-.....((((((.(((((.(((.....))).(((((.......))))))))))(((((.......))))))))))) ( -28.90, z-score = -1.58, R) >droGri2.scaffold_14906 12588238 74 + 14172833 ----------------------------------UGUGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAA ----------------------------------....((((((((((...((((.........))))...)).)))))))).(((((((.......))))).))... ( -27.90, z-score = -1.25, R) >anoGam1.chr3R 39615342 105 - 53272125 ---GCUGCUAAUUAUUUUGCACUCCCGUACACUAUCUGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCGAUCCCGGGCGGAAA ---..(((..........)))...(((((((((((........)))))))).)))......(.(((((.......))))).).(((((((.......))))).))... ( -29.80, z-score = 0.01, R) >consensus ______________GUUGUGAAUCCACCCCAUCACGUGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAA ......................................((((((((((...((((.........))))...)).)))))))).(((((((.......))))).))... (-27.51 = -27.46 + -0.05)
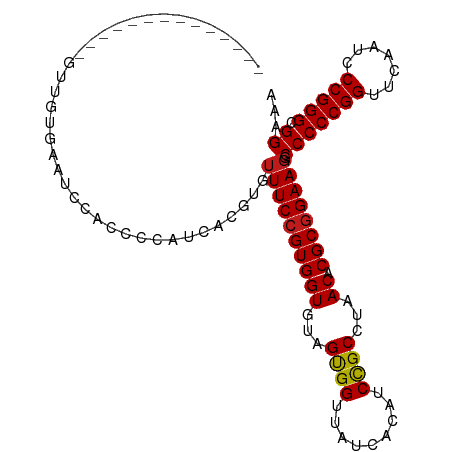
| Location | 12,147,388 – 12,147,482 |
|---|---|
| Length | 94 |
| Sequences | 13 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.88 |
| Shannon entropy | 0.46120 |
| G+C content | 0.56976 |
| Mean single sequence MFE | -34.72 |
| Consensus MFE | -26.79 |
| Energy contribution | -26.52 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.969353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12147388 94 - 27905053 UUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACACGUGAUGGGGUGGAUUCACAGC-------------- ..(((.(((((.......))))).((((..(....)..)))))))(((((...((((..((.((((......)))).))..))))..)))))..-------------- ( -40.70, z-score = -2.76, R) >droSim1.chr3R 18200150 94 + 27517382 UUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACACGUGAUGGGGUGGAUUCACAGC-------------- ..(((.(((((.......))))).((((..(....)..)))))))(((((...((((..((.((((......)))).))..))))..)))))..-------------- ( -40.70, z-score = -2.76, R) >droSec1.super_12 123702 94 + 2123299 UUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACACGUGAUGGGGUGGAUUCACAGC-------------- ..(((.(((((.......))))).((((..(....)..)))))))(((((...((((..((.((((......)))).))..))))..)))))..-------------- ( -40.70, z-score = -2.76, R) >droYak2.chr3R 14562336 94 + 28832112 UUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACACGUGAUGGGGUGGAUUUACAAC-------------- ..(((.(((((.......))))).((((..(....)..)))))))(((((...((((..((.((((......)))).))..))))..)))))..-------------- ( -38.50, z-score = -2.56, R) >droEre2.scaffold_4770 11672693 94 - 17746568 UUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACACGUGAUGGGGUUGAUUCACAAC-------------- ..(((.(((((.......))))).((((..(....)..)))))))((((((((((....((.((((......)))).)).)))))..)))))..-------------- ( -36.30, z-score = -1.93, R) >droWil1.scaffold_180708 3501503 106 - 12563649 UUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGCU-UGGGCGAUGCC-GGCCAUAAUGCAAUGACAAG (((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))..((.-(((.((....)-).)))....))......... ( -33.30, z-score = 0.18, R) >droPer1.super_3 6521062 102 - 7375914 UUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACUGCGAUGGACACUGCCUUGCCGAAAGGCAAC------ ......(((((.......)))))(((..(((((((....((.(...(((.....)))..).))..(....).))).))))....)))(((((....))))).------ ( -37.00, z-score = -1.57, R) >dp4.chr2 8330741 94 + 30794189 UUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCGCUACACCACGGAAACAUCC-UCGAGGGUGGC-CACAAAAA------------ ......(((((.......)))))(((((((((.((((..(((((.........)))))))))..)))))..((((-....)))))))-).......------------ ( -37.20, z-score = -1.41, R) >droAna3.scaffold_13340 15499175 93 - 23697760 UUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACUCUUGAGAGGGUUGCUUUCAAA--------------- (((((((((((.......)))))((...(((((.......))))).(((.....)))....)).))))))...(((((((.....))))))).--------------- ( -32.30, z-score = -0.99, R) >droVir3.scaffold_13047 1772924 87 - 19223366 UUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACACAAUGAAUACAGAGA--------------------- (((((((((((.......)))))((...(((((.......))))).(((.....)))....)).)))))).................--------------------- ( -26.70, z-score = -1.03, R) >droMoj3.scaffold_6540 18740883 80 + 34148556 UUUCCGCCCGGGAUUGAACCGGGGGCGUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACUUGG-UCAAA--------------------------- (((((((((((.......)))))((.(((((((.......))))).(((.....))).)).)).)))))).....-.....--------------------------- ( -28.20, z-score = -0.73, R) >droGri2.scaffold_14906 12588238 74 - 14172833 UUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACACA---------------------------------- (((((((((((.......)))))((...(((((.......))))).(((.....)))....)).))))))....---------------------------------- ( -26.70, z-score = -1.24, R) >anoGam1.chr3R 39615342 105 + 53272125 UUUCCGCCCGGGAUCGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACAGAUAGUGUACGGGAGUGCAAAAUAAUUAGCAGC--- (((((((((((.......)))))((...(((((.......))))).(((.....)))....)).)))))).......(((((....)))))..............--- ( -33.10, z-score = -0.65, R) >consensus UUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACACGUGAUGGGGUGGAUUCACAAC______________ (((((((((((.......)))))((...(((((.......))))).(((.....)))....)).))))))...................................... (-26.79 = -26.52 + -0.27)

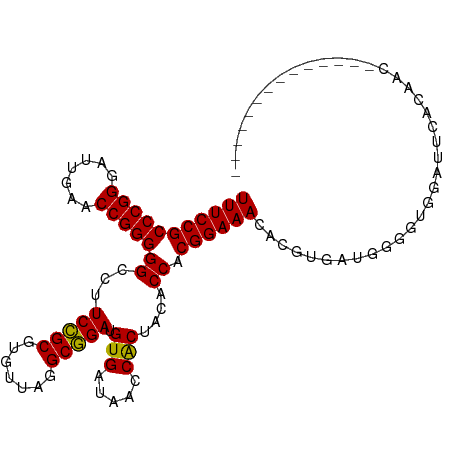
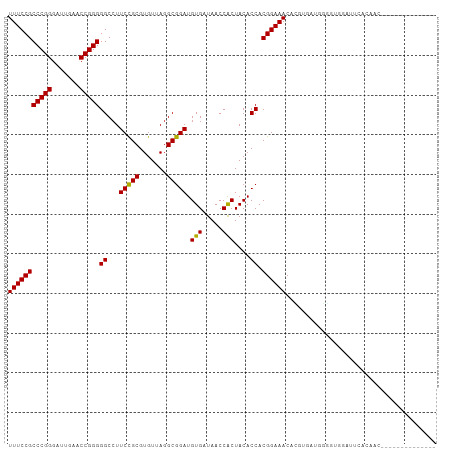
| Location | 12,147,407 – 12,147,500 |
|---|---|
| Length | 93 |
| Sequences | 13 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 82.59 |
| Shannon entropy | 0.38289 |
| G+C content | 0.51590 |
| Mean single sequence MFE | -32.32 |
| Consensus MFE | -29.60 |
| Energy contribution | -29.45 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.983683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12147407 93 + 27905053 -----ACGUGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAACAUUGG-AAAUAUUUAUUU---------- -----..((((((((((((((.......)))))).(((((.......))))).(.(((((((.......))))).))...)...))-))))))......---------- ( -32.00, z-score = -1.58, R) >droSim1.chr3R 18200169 94 - 27517382 -----ACGUGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAACAUAGGGAAAUAUUUAUUU---------- -----..(((((((((.(((.(.(((.....))).(((((.......)))))).)))(((((.......))))))))))))))................---------- ( -32.20, z-score = -1.39, R) >droSec1.super_12 123721 94 - 2123299 -----ACGUGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAACAUAGGAAAAUAUUUAUUU---------- -----..(((((((((.(((.(.(((.....))).(((((.......)))))).)))(((((.......))))))))))))))................---------- ( -32.20, z-score = -1.82, R) >droYak2.chr3R 14562355 93 - 28832112 -----ACGUGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAACAUUGC-AAUUAUUUAUUU---------- -----..(((((((((.(((.(.(((.....))).(((((.......)))))).)))(((((.......))))))))))))))...-............---------- ( -31.60, z-score = -1.83, R) >droEre2.scaffold_4770 11672712 88 + 17746568 -----ACGUGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAACAUUUUAACAUUU---------------- -----..(((((((((.(((.(.(((.....))).(((((.......)))))).)))(((((.......))))))))))))))..........---------------- ( -31.60, z-score = -2.15, R) >droWil1.scaffold_180708 3501534 93 + 12563649 -----AGCUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAUGUGAACUUCUUUUUU----------- -----.(((((((((((((((.......)))))).(((((.......)))))..((((..((......)).))))))))))).)).............----------- ( -29.70, z-score = -1.19, R) >droPer1.super_3 6521092 95 + 7375914 --------AGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAACA-GUUGAAAUAUUUAUUUUUACA----- --------.(((((((.(((.(.(((.....))).(((((.......)))))).)))(((((.......)))))))))))).-((.(((((....))))).)).----- ( -31.60, z-score = -2.22, R) >dp4.chr2 8330760 99 - 30794189 -----GGAUGUUUCCGUGGUGUAGCGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAACAUGGGGAAAUCUUUUUUUUUCUC----- -----..((((((((((((((.......)))))).(((((.......)))))..((((..((......)).))))))))))))(((((((.......)))))))----- ( -35.00, z-score = -1.37, R) >droAna3.scaffold_13340 15499193 97 + 23697760 -----AAGAGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAACAUUUGGGUUUCUUUUUUAAUU------- -----(((((((((((((((...((((.........))))...)).))))))))((((((((.......))))..(....)....)))))))))........------- ( -30.50, z-score = -1.07, R) >droVir3.scaffold_13047 1772937 93 + 19223366 ------UGUGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAACAUCAUCUAAUCAAGACUA---------- ------.(((((((((.(((.(.(((.....))).(((((.......)))))).)))(((((.......))))))))))))))................---------- ( -31.50, z-score = -1.89, R) >droMoj3.scaffold_6540 18740889 81 - 34148556 ------CAAGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAACGCCCCCGGUUCAAUCCCGGGCGGAAACAUAUUG---------------------- ------...(((((((.(((((.(((.....))).(((((.......))))))))))(((((.......))))))))))))......---------------------- ( -33.30, z-score = -3.62, R) >droGri2.scaffold_14906 12588238 103 + 14172833 ------UGUGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAACAUUCGGCCCAAUCCUCAUUUGGAGUUUU ------.....(((((((((...((((.........))))...)).)))))))(((((((((.......))))).(....)....))))...(((......)))..... ( -36.10, z-score = -1.43, R) >anoGam1.chr3R 39615367 103 - 53272125 ACACUAUCUGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAUGUGAUUUUUUUUCUGUAUUA------ .(((....((((((((.(((.(.(((.....))).(((((.......)))))).)))(((((.......))))))))))))).))).................------ ( -32.90, z-score = -1.67, R) >consensus _____ACGUGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAACAUUUGGAAAUAUUUAUUU__________ .........((((((((((((.......)))))).(((((.......)))))..((((..((......)).))))))))))............................ (-29.60 = -29.45 + -0.14)

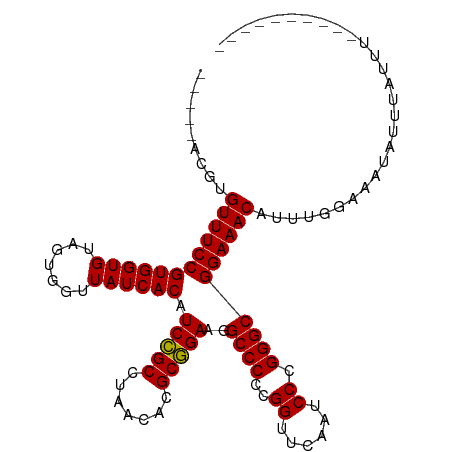
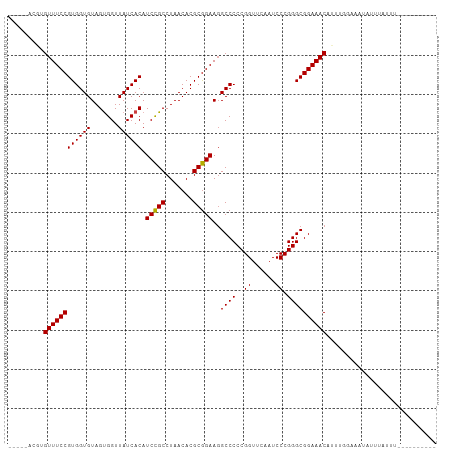
| Location | 12,147,407 – 12,147,500 |
|---|---|
| Length | 93 |
| Sequences | 13 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 82.59 |
| Shannon entropy | 0.38289 |
| G+C content | 0.51590 |
| Mean single sequence MFE | -31.50 |
| Consensus MFE | -30.58 |
| Energy contribution | -30.31 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.979141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12147407 93 - 27905053 ----------AAAUAAAUAUUU-CCAAUGUUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACACGU----- ----------............-....(((((((((((((.......)))))((...(((((.......))))).(((.....)))....)).))))))))...----- ( -31.30, z-score = -1.67, R) >droSim1.chr3R 18200169 94 + 27517382 ----------AAAUAAAUAUUUCCCUAUGUUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACACGU----- ----------.................(((((((((((((.......)))))((...(((((.......))))).(((.....)))....)).))))))))...----- ( -31.30, z-score = -1.51, R) >droSec1.super_12 123721 94 + 2123299 ----------AAAUAAAUAUUUUCCUAUGUUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACACGU----- ----------.................(((((((((((((.......)))))((...(((((.......))))).(((.....)))....)).))))))))...----- ( -31.30, z-score = -1.69, R) >droYak2.chr3R 14562355 93 + 28832112 ----------AAAUAAAUAAUU-GCAAUGUUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACACGU----- ----------............-....(((((((((((((.......)))))((...(((((.......))))).(((.....)))....)).))))))))...----- ( -31.30, z-score = -1.66, R) >droEre2.scaffold_4770 11672712 88 - 17746568 ----------------AAAUGUUAAAAUGUUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACACGU----- ----------------...........(((((((((((((.......)))))((...(((((.......))))).(((.....)))....)).))))))))...----- ( -31.30, z-score = -1.66, R) >droWil1.scaffold_180708 3501534 93 - 12563649 -----------AAAAAAGAAGUUCACAUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGCU----- -----------................(((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))...----- ( -29.70, z-score = -1.26, R) >droPer1.super_3 6521092 95 - 7375914 -----UGUAAAAAUAAAUAUUUCAAC-UGUUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACU-------- -----.....................-.((((((((((((.......)))))((...(((((.......))))).(((.....)))....)).))))))).-------- ( -30.10, z-score = -1.71, R) >dp4.chr2 8330760 99 + 30794189 -----GAGAAAAAAAAAGAUUUCCCCAUGUUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCGCUACACCACGGAAACAUCC----- -----.....................((((((((((((((.......)))))........(.((((..(((((.........)))))))))).)))))))))..----- ( -31.90, z-score = -1.17, R) >droAna3.scaffold_13340 15499193 97 - 23697760 -------AAUUAAAAAAGAAACCCAAAUGUUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACUCUU----- -------.....................((((((((((((.......)))))((...(((((.......))))).(((.....)))....)).)))))))....----- ( -30.10, z-score = -1.45, R) >droVir3.scaffold_13047 1772937 93 - 19223366 ----------UAGUCUUGAUUAGAUGAUGUUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACACA------ ----------..((((.....))))..(((((((((((((.......)))))((...(((((.......))))).(((.....)))....)).))))))))..------ ( -32.90, z-score = -1.97, R) >droMoj3.scaffold_6540 18740889 81 + 34148556 ----------------------CAAUAUGUUUCCGCCCGGGAUUGAACCGGGGGCGUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACUUG------ ----------------------......((((((((((((.......)))))((.(((((((.......))))).(((.....))).)).)).)))))))...------ ( -31.60, z-score = -2.38, R) >droGri2.scaffold_14906 12588238 103 - 14172833 AAAACUCCAAAUGAGGAUUGGGCCGAAUGUUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACACA------ .....(((((.......))))).....(((((((((((((.......)))))((...(((((.......))))).(((.....)))....)).))))))))..------ ( -33.60, z-score = -0.51, R) >anoGam1.chr3R 39615367 103 + 53272125 ------UAAUACAGAAAAAAAAUCACAUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACAGAUAGUGU ------.................(((.(((((((((((((.......)))))((...(((((.......))))).(((.....)))....)).))))))))....))). ( -33.10, z-score = -2.05, R) >consensus __________AAAUAAAUAUUUCCAAAUGUUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACACGU_____ ............................((((((((((((.......)))))((...(((((.......))))).(((.....)))....)).)))))))......... (-30.58 = -30.31 + -0.27)

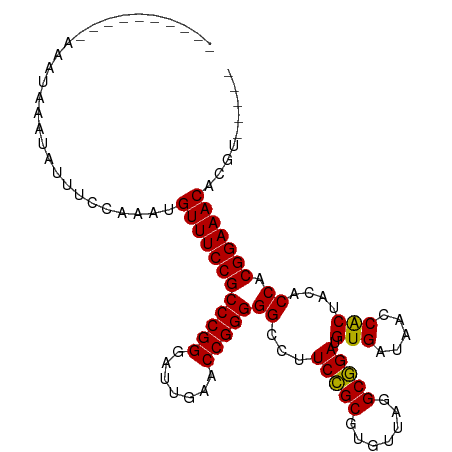
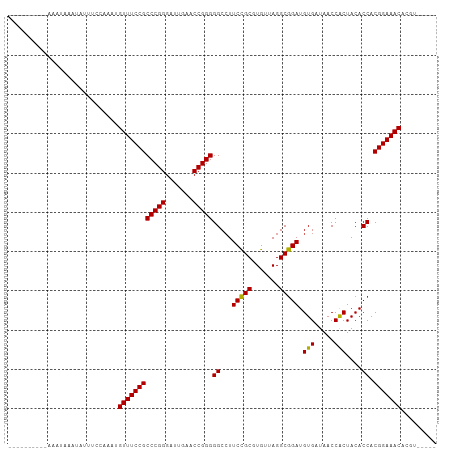
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:19:35 2011