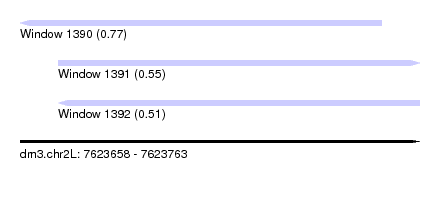
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,623,658 – 7,623,763 |
| Length | 105 |
| Max. P | 0.772583 |

| Location | 7,623,658 – 7,623,753 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 77.09 |
| Shannon entropy | 0.38729 |
| G+C content | 0.34608 |
| Mean single sequence MFE | -18.88 |
| Consensus MFE | -8.74 |
| Energy contribution | -9.14 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.772583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
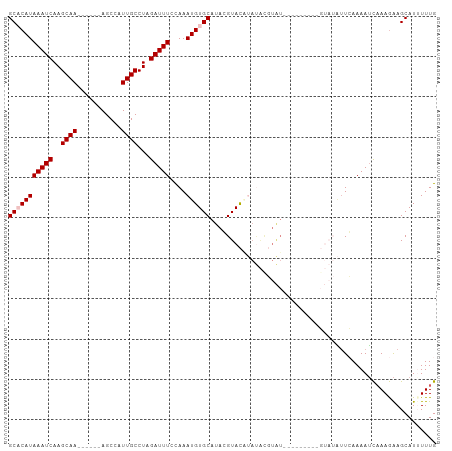
>dm3.chr2L 7623658 95 - 23011544 GCACAUAAAUCAAGCAA------AGCCAUUGCCUAGAUUUCCAAAUGUGCAUACGUACCUAUAUGUAUGUA-----CGUAUAUUCAAAAUCAAAGAAGCAUUUUUG (((((((((((..((((------.....))))...)))))....))))))((((((((.((....)).)))-----))))).........((((((....)))))) ( -21.10, z-score = -3.33, R) >droSim1.chr2L 7420575 91 - 22036055 GCCCAUAAAUCAAGCAA------AGCCAUUGCCUAGAUUUCCAAAUGUGCAUAGGUACGUAUACGUAU---------GUAUAUUCAAAAUCAAAGAAGCAUAUUUG ......(((((..((((------.....))))...))))).((((((((((((.(((((....)))))---------.))).(((.........)))))))))))) ( -20.30, z-score = -3.24, R) >droSec1.super_3 3137363 91 - 7220098 GCCCAUAAAUCAAGCAA------AGCCAUUGCCUAGAUUUCCAAAUGUGCAUAGGUACAUAUACGUAU---------GUAUAUUCAAAAUCAAAGAAGCAUAUUUG ((...........((((------.....))))...(((((...((((((((((.(((....))).)))---------)))))))..)))))......))....... ( -18.50, z-score = -2.65, R) >droYak2.chr2L 17053035 78 + 22324452 GCACAUAAAUCAAGCAA------AGCCAUUGCCUAGAUUUCUAAAUGUGCAUACGUAU----------------------AGUUGAAAAUCAAAGAAGCAUUUUUG (((((((((((..((((------.....))))...)))))....))))))....((.(----------------------..(((.....)))..).))....... ( -13.90, z-score = -1.17, R) >droEre2.scaffold_4929 16543066 106 - 26641161 GCACAUAAAUCAAGCAAAGCCAUAGCCAUUGCCUAGAUUUCUAAAUGUGCAUACGUACACACGUCUGUGUAUUCAUUCGCAUAUGAAAAAUCAAAGAGCAUUUUUG (((((((((((..((((.((....))..))))...)))))....))))))....(((((((....)))))))((((......)))).................... ( -20.60, z-score = -1.20, R) >consensus GCACAUAAAUCAAGCAA______AGCCAUUGCCUAGAUUUCCAAAUGUGCAUACGUACAUAUACGUAU_________GUAUAUUCAAAAUCAAAGAAGCAUUUUUG (((((((((((..((((...........))))...)))))....))))))........................................................ ( -8.74 = -9.14 + 0.40)

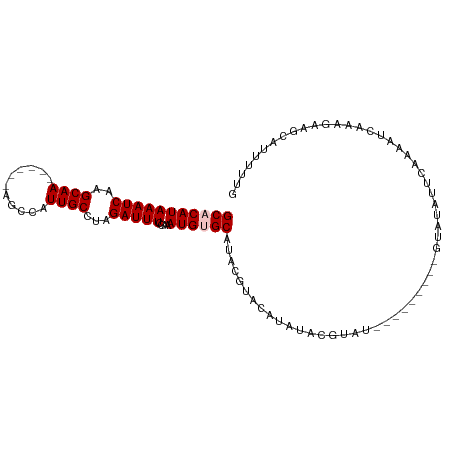
| Location | 7,623,668 – 7,623,763 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.14 |
| Shannon entropy | 0.31471 |
| G+C content | 0.37820 |
| Mean single sequence MFE | -23.52 |
| Consensus MFE | -13.62 |
| Energy contribution | -13.62 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.548152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7623668 95 + 23011544 UCUUUGAUUUUGAAUAUACG------UACAUACAUAUAGGUACGUAUGCACAUUUGGAAAUCUAGGCAAUGGCU------UUGCUUGAUUUAUGUGCUUGUGCCCAC ..............((((.(------(....)).))))((((((...((((((....(((((.((((((.....------))))))))))))))))).))))))... ( -24.20, z-score = -1.84, R) >droSim1.chr2L 7420585 91 + 22036055 UCUUUGAUUUUGAAUA----------UACAUACGUAUACGUACCUAUGCACAUUUGGAAAUCUAGGCAAUGGCU------UUGCUUGAUUUAUGGGCUUGUGCCCAC .....((((((.((..----------..((((.(((....))).)))).....)).))))))(((((((.....------))))))).....(((((....))))). ( -22.90, z-score = -2.15, R) >droSec1.super_3 3137373 91 + 7220098 UCUUUGAUUUUGAAUA----------UACAUACGUAUAUGUACCUAUGCACAUUUGGAAAUCUAGGCAAUGGCU------UUGCUUGAUUUAUGGGCUUGUGCCCAC .....((((((.((..----------..((((.(((....))).)))).....)).))))))(((((((.....------))))))).....(((((....))))). ( -22.90, z-score = -1.99, R) >droYak2.chr2L 17053045 78 - 22324452 UCUUUGAUUUUCAAC-------------------UAUACG----UAUGCACAUUUAGAAAUCUAGGCAAUGGCU------UUGCUUGAUUUAUGUGCUUGUGCCCAC ...............-------------------..((((----...((((((....(((((.((((((.....------))))))))))))))))).))))..... ( -17.40, z-score = -1.55, R) >droEre2.scaffold_4929 16543075 107 + 26641161 UCUUUGAUUUUUCAUAUGCGAAUGAAUACACAGACGUGUGUACGUAUGCACAUUUAGAAAUCUAGGCAAUGGCUAUGGCUUUGCUUGAUUUAUGUGCUUGUGCCCAC (((.((....(((((......)))))..)).))).(((.(((((...((((((....(((((.((((((.(((....)))))))))))))))))))).))))).))) ( -30.20, z-score = -2.15, R) >consensus UCUUUGAUUUUGAAUA__________UACAUACAUAUACGUACCUAUGCACAUUUGGAAAUCUAGGCAAUGGCU______UUGCUUGAUUUAUGUGCUUGUGCCCAC ...............................................(((((.....(((((.((((((...........))))))))))).......))))).... (-13.62 = -13.62 + 0.00)



| Location | 7,623,668 – 7,623,763 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 81.14 |
| Shannon entropy | 0.31471 |
| G+C content | 0.37820 |
| Mean single sequence MFE | -22.86 |
| Consensus MFE | -9.26 |
| Energy contribution | -9.26 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.508932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7623668 95 - 23011544 GUGGGCACAAGCACAUAAAUCAAGCAA------AGCCAUUGCCUAGAUUUCCAAAUGUGCAUACGUACCUAUAUGUAUGUA------CGUAUAUUCAAAAUCAAAGA .((((((...((...............------.))...))))))(((((...(((((((.(((((((......)))))))------.)))))))..)))))..... ( -23.09, z-score = -2.65, R) >droSim1.chr2L 7420585 91 - 22036055 GUGGGCACAAGCCCAUAAAUCAAGCAA------AGCCAUUGCCUAGAUUUCCAAAUGUGCAUAGGUACGUAUACGUAUGUA----------UAUUCAAAAUCAAAGA ((((((....)))))).......((((------.....))))...(((((...((((((((((.(((....))).))))))----------))))..)))))..... ( -26.60, z-score = -4.16, R) >droSec1.super_3 3137373 91 - 7220098 GUGGGCACAAGCCCAUAAAUCAAGCAA------AGCCAUUGCCUAGAUUUCCAAAUGUGCAUAGGUACAUAUACGUAUGUA----------UAUUCAAAAUCAAAGA ((((((....)))))).......((((------.....))))...(((((...((((((((((.(((....))).))))))----------))))..)))))..... ( -26.60, z-score = -4.32, R) >droYak2.chr2L 17053045 78 + 22324452 GUGGGCACAAGCACAUAAAUCAAGCAA------AGCCAUUGCCUAGAUUUCUAAAUGUGCAUA----CGUAUA-------------------GUUGAAAAUCAAAGA .((((((...((...............------.))...))))))(((((.(((.(((((...----.)))))-------------------.))).)))))..... ( -15.09, z-score = -0.81, R) >droEre2.scaffold_4929 16543075 107 - 26641161 GUGGGCACAAGCACAUAAAUCAAGCAAAGCCAUAGCCAUUGCCUAGAUUUCUAAAUGUGCAUACGUACACACGUCUGUGUAUUCAUUCGCAUAUGAAAAAUCAAAGA .((((((...((...........))...((....))...))))))(((((....((((((....(((((((....)))))))......))))))...)))))..... ( -22.90, z-score = -1.03, R) >consensus GUGGGCACAAGCACAUAAAUCAAGCAA______AGCCAUUGCCUAGAUUUCCAAAUGUGCAUACGUACAUAUACGUAUGUA__________UAUUCAAAAUCAAAGA ....(((((.......(((((..((((...........))))...))))).....)))))............................................... ( -9.26 = -9.26 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:24:03 2011