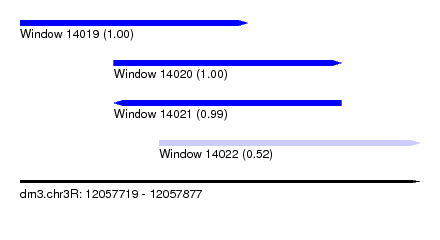
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,057,719 – 12,057,877 |
| Length | 158 |
| Max. P | 0.998490 |

| Location | 12,057,719 – 12,057,809 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 87.31 |
| Shannon entropy | 0.17346 |
| G+C content | 0.43241 |
| Mean single sequence MFE | -25.70 |
| Consensus MFE | -22.43 |
| Energy contribution | -22.10 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.11 |
| SVM RNA-class probability | 0.997466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12057719 90 + 27905053 GUCUCUUUUCUCGACUUCGUCAAACUCGUUUGAACUGCGCAAGUUAGGUUAUGGCUCGGGGAUUUCCCCUAGAAUAUAACUAAUUUGUGU (((.........)))....(((((....)))))...(((((((((((.(((((.((.((((....)))).))..)))))))))))))))) ( -26.50, z-score = -3.20, R) >droSim1.chr3R 18108921 88 - 27517382 --GUCUUCUCUCGACUUCGUCAAACUCGUUAGCAUCGCGCAAGUUAAGUUAUUGCUGAGGGACUUCCCCUCAAAUAUAACUAAUUUGUGU --(((.......))).....................((((((((((.(((((...((((((.....))))))...))))))))))))))) ( -24.90, z-score = -4.10, R) >droSec1.super_219 16512 88 - 23606 --GUCUUCUCUCGACUUCGUCAAGCUCGUUUGAACCGCGCAAGUUAAGUUAUUGCUGGGGGACUUCCCCUCAAACAUAACUAAUUUGUGU --(((.......)))....(((((....)))))...((((((((((.(((((...((((((.....))))))...))))))))))))))) ( -25.70, z-score = -3.16, R) >consensus __GUCUUCUCUCGACUUCGUCAAACUCGUUUGAACCGCGCAAGUUAAGUUAUUGCUGGGGGACUUCCCCUCAAAUAUAACUAAUUUGUGU ............(((...)))...............(((((((((.((((((...((((((.....))))))...))))))))))))))) (-22.43 = -22.10 + -0.33)

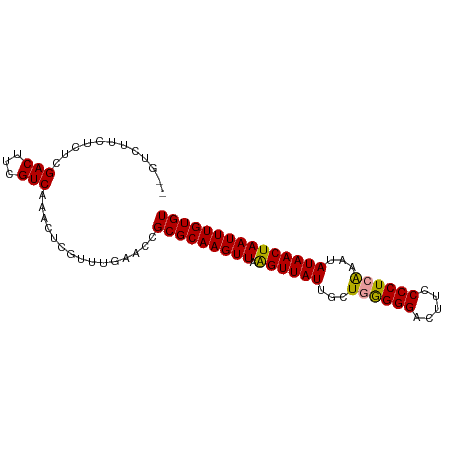
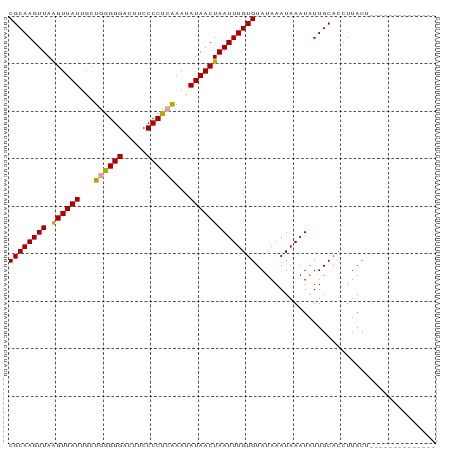
| Location | 12,057,756 – 12,057,846 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 81.25 |
| Shannon entropy | 0.24488 |
| G+C content | 0.35029 |
| Mean single sequence MFE | -21.47 |
| Consensus MFE | -19.86 |
| Energy contribution | -19.53 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.38 |
| SVM RNA-class probability | 0.998490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12057756 90 + 27905053 CGCAAGUUAGGUUAUGGCUCGGGGAUUUCCCCUAGAAUAUAACUAAUUUGUGUAUAAAUAAAUGUUGCACCUUACCAAAACUAACUGACU .(..((((((((((((.((.((((....)))).))..))))))......(((((...........)))))..........))))))..). ( -22.20, z-score = -2.26, R) >droSim1.chr3R 18108956 76 - 27517382 CGCAAGUUAAGUUAUUGCUGAGGGACUUCCCCUCAAAUAUAACUAAUUUGUGUAUAAAUAAAUAUUGCACCUUACU-------------- (((((((((.(((((...((((((.....))))))...))))))))))))))........................-------------- ( -21.90, z-score = -4.78, R) >droSec1.super_219 16547 76 - 23606 CGCAAGUUAAGUUAUUGCUGGGGGACUUCCCCUCAAACAUAACUAAUUUGUGUAUAAAUAAAUAUUGCACCUUACU-------------- (((((((((.(((((...((((((.....))))))...))))))))))))))........................-------------- ( -20.30, z-score = -3.20, R) >consensus CGCAAGUUAAGUUAUUGCUGGGGGACUUCCCCUCAAAUAUAACUAAUUUGUGUAUAAAUAAAUAUUGCACCUUACU______________ ((((((((.((((((...((((((.....))))))...))))))))))))))...................................... (-19.86 = -19.53 + -0.33)

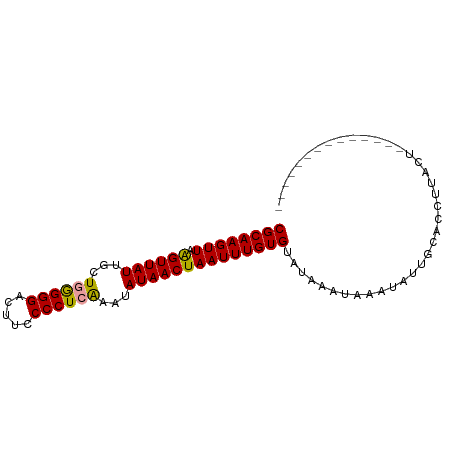
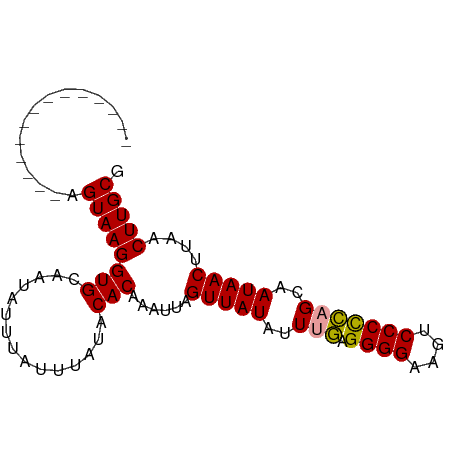
| Location | 12,057,756 – 12,057,846 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 81.25 |
| Shannon entropy | 0.24488 |
| G+C content | 0.35029 |
| Mean single sequence MFE | -20.75 |
| Consensus MFE | -16.19 |
| Energy contribution | -15.86 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.989515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12057756 90 - 27905053 AGUCAGUUAGUUUUGGUAAGGUGCAACAUUUAUUUAUACACAAAUUAGUUAUAUUCUAGGGGAAAUCCCCGAGCCAUAACCUAACUUGCG .((.((((((((.((((...(((...............))).................((((....))))..)))).)).)))))).)). ( -21.36, z-score = -2.07, R) >droSim1.chr3R 18108956 76 + 27517382 --------------AGUAAGGUGCAAUAUUUAUUUAUACACAAAUUAGUUAUAUUUGAGGGGAAGUCCCUCAGCAAUAACUUAACUUGCG --------------.((((((((..(((......))).))).....((((((..(((((((.....)))))))..))))))...))))). ( -20.50, z-score = -3.62, R) >droSec1.super_219 16547 76 + 23606 --------------AGUAAGGUGCAAUAUUUAUUUAUACACAAAUUAGUUAUGUUUGAGGGGAAGUCCCCCAGCAAUAACUUAACUUGCG --------------.((((((((..(((......))).))).....((((((..(((.((((....)))))))..))))))...))))). ( -20.40, z-score = -2.82, R) >consensus ______________AGUAAGGUGCAAUAUUUAUUUAUACACAAAUUAGUUAUAUUUGAGGGGAAGUCCCCCAGCAAUAACUUAACUUGCG ...............((((((((...............)))......(((((..(((.((((....)))))))..)))))....))))). (-16.19 = -15.86 + -0.33)

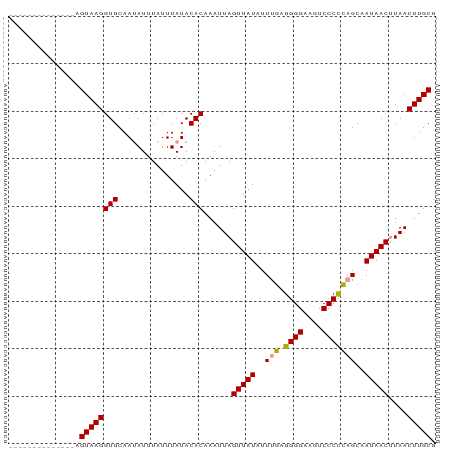
| Location | 12,057,774 – 12,057,877 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 80.14 |
| Shannon entropy | 0.25855 |
| G+C content | 0.32321 |
| Mean single sequence MFE | -15.20 |
| Consensus MFE | -10.81 |
| Energy contribution | -10.70 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.522368 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 12057774 103 + 27905053 UCGGGGAUUUCCCCUAGAAUAUAACUAAUUUGUGUAUAAAUAAAUGUUGCACCUUACCAAAACUAACUGACUGCCAUAAACACCCAUAUUCGUUUUAUAAUAC ..((((....))))..((((((.........(((((...........))))).....((........))................))))))............ ( -14.00, z-score = -0.48, R) >droSim1.chr3R 18108974 86 - 27517382 UGAGGGACUUCCCCUCAAAUAUAACUAAUUUGUGUAUAAAUAAAUAUUGCACCUUAC--------------UGCCAUAAAUACACAUCUUCGUUUUAUAU--- ((((((.....)))))).(((.(((.((..(((((((.......(((.(((......--------------))).))).)))))))..)).))).)))..--- ( -18.20, z-score = -3.84, R) >droSec1.super_219 16565 86 - 23606 UGGGGGACUUCCCCUCAAACAUAACUAAUUUGUGUAUAAAUAAAUAUUGCACCUUAC--------------UGCCAUAAAUACCCAUCAUCGUUUUAUAU--- ((((((.....)))))).((((((.....)))))).........(((.(((......--------------))).)))......................--- ( -13.40, z-score = -0.70, R) >consensus UGGGGGACUUCCCCUCAAAUAUAACUAAUUUGUGUAUAAAUAAAUAUUGCACCUUAC______________UGCCAUAAAUACCCAUCUUCGUUUUAUAU___ ((((((.....))))))..............(((((...........)))))................................................... (-10.81 = -10.70 + -0.11)

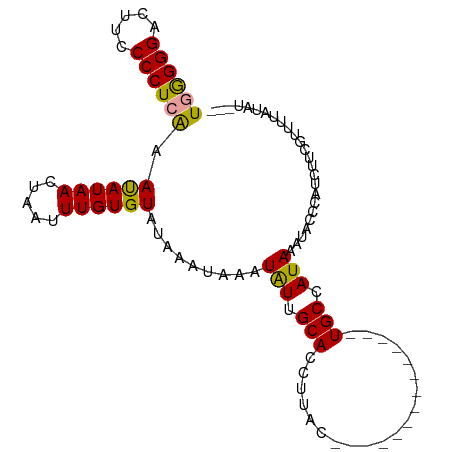
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:19:19 2011