
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,044,108 – 12,044,200 |
| Length | 92 |
| Max. P | 0.744832 |

| Location | 12,044,108 – 12,044,200 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 71.60 |
| Shannon entropy | 0.54103 |
| G+C content | 0.57665 |
| Mean single sequence MFE | -38.88 |
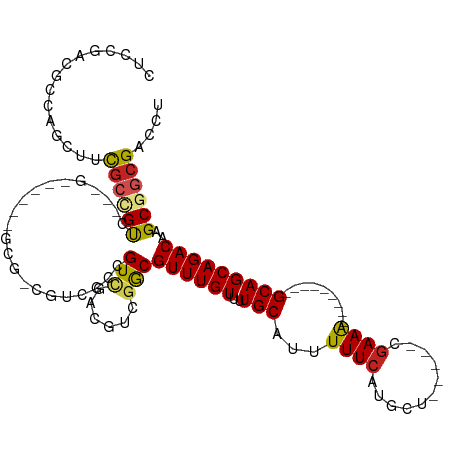
| Consensus MFE | -15.00 |
| Energy contribution | -14.91 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.744832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
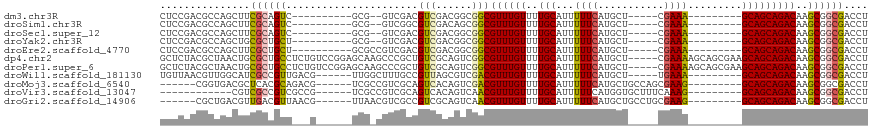
>dm3.chr3R 12044108 92 + 27905053 CUCCGACGCCAGCUUCGCAGUC----------GCG--GUCGACGUCGACGGCGGCGUUUGUUUUGCAUUUUUCAUGCU-----CGAAA---------GCAGCAGACAAGCGGCGACCU ......((((.........(((----------((.--((((....)))).)))))((((((((.((..(((((.....-----.))))---------)..)))))))))))))).... ( -37.20, z-score = -1.59, R) >droSim1.chr3R 18085148 92 - 27517382 CUCCGACGCCAGCUUCGCAGUC----------GCG--GUCGGCGUCGACAGCGGCGUUUGUUUUGCAUUUUUCAUGCU-----CGAAA---------GCAGCAGACAAGCGGCGACCU ...(((((((.((..(((....----------)))--)).)))))))....((.(((((((((.((..(((((.....-----.))))---------)..))))))))))).)).... ( -36.50, z-score = -1.43, R) >droSec1.super_12 18701 92 - 2123299 CUCCGACGCCAGCUUCGCAGUC----------GCG--GUCGACGUCGACGGCGGCGUUUGUUUUGCAUUUUUCAUGCU-----CGAAA---------GCAGCAGACAAGCGGCGACCU ......((((.........(((----------((.--((((....)))).)))))((((((((.((..(((((.....-----.))))---------)..)))))))))))))).... ( -37.20, z-score = -1.59, R) >droYak2.chr3R 14444831 92 - 28832112 CUCCGACGCCAGCUGCGCUGCU----------GCG--GUCGACGUCGACGGCGGCGUUUGUUUUGCAUUUUUCAUGCU-----CGAAA---------GCAGCAGACAAGCGGCGACCU ......((((.(((...(((((----------((.--.(((((((((....)))))........((((.....)))))-----)))..---------)))))))...))))))).... ( -40.30, z-score = -1.81, R) >droEre2.scaffold_4770 11567743 94 + 17746568 CUCCGACGCCAGCUUCGCUGCU----------GCGCCGUCGACGUCGACGGCGGCGUUUGUUUUGCAUUUUUCAUGCU-----CGAAA---------GCAGCAGACAAGCGGCGACCU ......((((.((((..(((((----------(((((((((....)))))))....((((....((((.....)))).-----)))).---------)))))))..)))))))).... ( -44.40, z-score = -3.25, R) >dp4.chr2 25289031 113 + 30794189 GCUCUACGCUAACUGCGCUGCCUCUGUCCGGAGCAAGCCCGCUGUCGCAGUCGGCGUUUGUUUUGCAUUUUUCAUGCU-----CGAAAAGCAGCGAAGCAGCAGACAAGCGGCGACCU ..((..((((..(((((((..(.((((.((((((((((((((((...))).))).))))))))))...(((((.....-----.))))))))).).))).))))...))))..))... ( -37.40, z-score = 0.82, R) >droPer1.super_6 617156 113 + 6141320 GCUCUACGCUAACUGCGCUGCCUCUGUCCGGAGCAAGCCCGCUGUCGCAGUCGGCGUUUGUUUUGCAUUUUUCAUGCU-----CGAAAAGCAGCGAAGCAGCAGACAAGCGGCGACCU ..((..((((..(((((((..(.((((.((((((((((((((((...))).))).))))))))))...(((((.....-----.))))))))).).))).))))...))))..))... ( -37.40, z-score = 0.82, R) >droWil1.scaffold_181130 15628254 98 + 16660200 UGUUAACGUUGGCAUCGCCGUUGACG------UUGGCUUUGCCGUUAGCGUCGACGUUUGUUUUGCAUUUUUCAUGCU-----UGAAA---------GCAGCAGACAAGCGGCGACCU (((((....)))))((((((((((((------(((((......))))))))))))((((((((.((..((((((....-----)))))---------)..)))))))))))))))... ( -41.10, z-score = -3.69, R) >droMoj3.scaffold_6540 13263809 97 - 34148556 ------CGGUGACGCUCACGCAGACG------UCGCCGUCGCAGUCACAGUCGACGUUUGUUUUGCAUUUUUCAUGCUGCCAGCGAAG---------GCAGCAGACAAGCGGCGACCU ------.(((((((.((.....))))------)))))(((((.(((......)))(((((((............(((((((......)---------))))))))))))).))))).. ( -42.10, z-score = -3.04, R) >droVir3.scaffold_13047 15343093 91 - 19223366 ------------CGUCGCCGUCGCCG------UCGCCGUCGCAGUCACAGUCAACGUUUGUUUUGCAUUUUUCAUGGUGCUUUCAAAG---------GCAGCAGACAAGCGGCGACCU ------------.((((((((.((..------..)).(((((((..((((.......)))).))))........((.(((((....))---------))).)))))..)))))))).. ( -30.30, z-score = -1.71, R) >droGri2.scaffold_14906 13638906 97 - 14172833 ------CGCUGACGUUGACGUUAACG------UUAACGUCGCCGUCGCAGUCAACGUUUGUUUUGCAUUUUUCAUGCUGCCUGCGAAG---------GCAGCAGACAAGCGGCGACCU ------.((.((((((((((....))------))))))))))((((((.(((...((.......))........((((((((....))---------)))))))))..)))))).... ( -43.80, z-score = -4.13, R) >consensus CUCCGACGCCAGCUUCGCCGUC___G______GCG_CGUCGCCGUCGACGUCGGCGUUUGUUUUGCAUUUUUCAUGCU_____CGAAA_________GCAGCAGACAAGCGGCGACCU ...............((((((......................(((......)))((((((..(((...............................)))))))))..)))))).... (-15.00 = -14.91 + -0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:19:15 2011