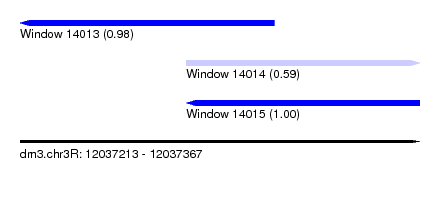
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,037,213 – 12,037,367 |
| Length | 154 |
| Max. P | 0.996462 |

| Location | 12,037,213 – 12,037,311 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 74.59 |
| Shannon entropy | 0.46333 |
| G+C content | 0.51937 |
| Mean single sequence MFE | -17.16 |
| Consensus MFE | -9.36 |
| Energy contribution | -11.24 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.979055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
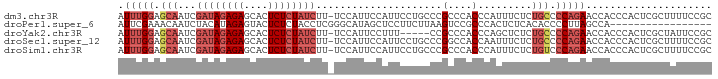
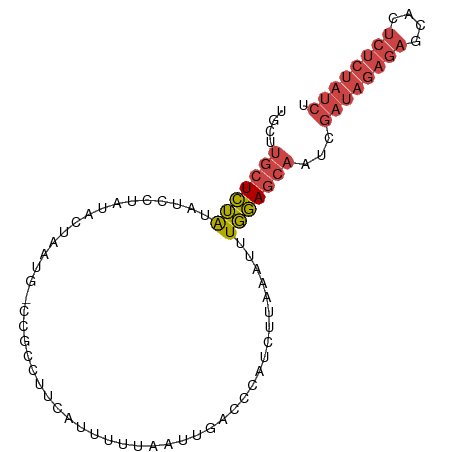
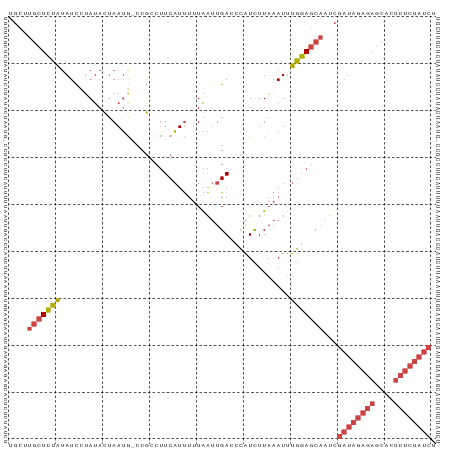
>dm3.chr3R 12037213 98 - 27905053 AUUUGGAGCAAUCGAUAGAGAGCACUCUCUAUCUU-UCCAUUCCAUUCCUGCCCGCCCACCCAUUUCUCUGCCCCAGAACCACCCACUCGCUUUUCCGC .(((((.(((...((((((((....))))))))..-..............(....).............))).)))))..................... ( -17.40, z-score = -2.46, R) >droPer1.super_6 610871 82 - 6141320 AUUCGAAACAAUCUACAUAGAGUACUCUCUACCUCGGGCAUAGCUCCUUCUUAAGUCCGCCCACUCUCACACCCCUUAGCCA----------------- ....((...........(((((....)))))....((((...(((........)))..))))....))..............----------------- ( -12.50, z-score = -1.61, R) >droYak2.chr3R 14438012 93 + 28832112 AUUUGGAGCAAUCGAUAGAGAGCACUCUCUAUCUU-UCCAUUCCUUU-----CCGCCCACCCAGCUCUCUGCCCCAGAACCACCCACUCGCUAUUCCGC .(((((.(((...((((((((....))))))))..-...........-----..((.......))....))).)))))..................... ( -19.80, z-score = -3.10, R) >droSec1.super_12 11885 98 + 2123299 AUUUGGAGCAAUCGAUAGAGAGCACUCUCUAUCUU-UCCAUUCCAUUCCUGCCCGGCCACCAAUUUCUCUGCCCCAGAACCACCCACUCGCUUUUCCGC .(((((.(((...((((((((....))))))))..-...........((.....)).............))).)))))..................... ( -19.40, z-score = -2.07, R) >droSim1.chr3R 18078370 98 + 27517382 AUUUGGAGCAAUCGAUAGAGAGCACUCUCUAUCUU-UCCAUUCCAUUCCUGCCCGCCCACCCAUUUCUCUGUCCCAGAACCACCCACUCGCUUUUCCGC ....(((((....((((((((....))))))))..-...............................((((...))))...........)))))..... ( -16.70, z-score = -2.15, R) >consensus AUUUGGAGCAAUCGAUAGAGAGCACUCUCUAUCUU_UCCAUUCCAUUCCUGCCCGCCCACCCAUUUCUCUGCCCCAGAACCACCCACUCGCUUUUCCGC .(((((.(((...((((((((....)))))))).....................(....).........))).)))))..................... ( -9.36 = -11.24 + 1.88)
| Location | 12,037,277 – 12,037,367 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 76.83 |
| Shannon entropy | 0.44302 |
| G+C content | 0.37579 |
| Mean single sequence MFE | -20.45 |
| Consensus MFE | -11.54 |
| Energy contribution | -12.30 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.589374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
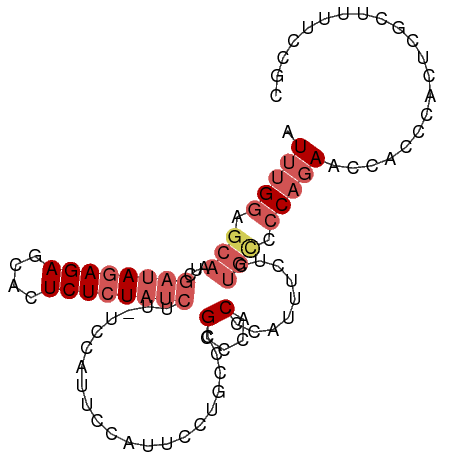
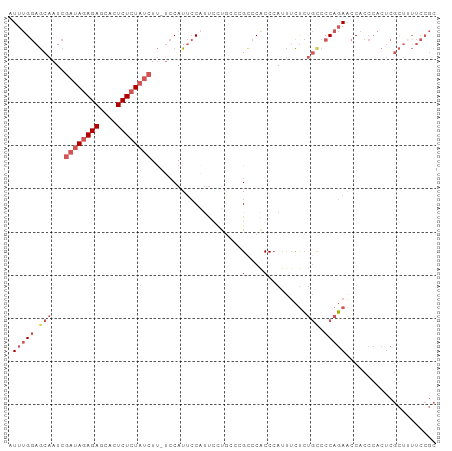
>dm3.chr3R 12037277 90 + 27905053 AGAUAGAGAGUGCUCUCUAUCGAUUGCUCCAAAUUUAAGAUGGGUCAAUUAAAAAUGAAGGCGG-CAUUAGUUUAGGAUAUAGAGCAAGCA .((((((((....))))))))(.((((((...(((..((((..(((................))-)....))))..)))...)))))).). ( -22.09, z-score = -1.46, R) >droAna3.scaffold_12911 2858539 82 - 5364042 -------GGAUAGAUUCGAAGGAUAGAUAGACAUAAAUAACCGAUCAGCCAGUGAAGAGGCUGAACUUUCAUAUUGAAUACCUAAAGUU-- -------((....((((((.((.((..((....))..)).))..((((((........)))))).........)))))).)).......-- ( -15.20, z-score = -1.41, R) >droEre2.scaffold_4770 11560924 90 + 17746568 AGAUAGAGAGUGCUCUCUAUCGAUUGCUCCAAAUUUAAGAUGGGUCAAUUAAAAAUGAAGGCGG-UAUUAGUAUAAAAUACAGAGCAGGCA .((((((((....))))))))(((((..(((.........)))..))))).....((...((.(-((((.......)))))...))...)) ( -22.10, z-score = -2.10, R) >droYak2.chr3R 14438071 90 - 28832112 AGAUAGAGAGUGCUCUCUAUCGAUUGCUCCAAAUUUAAGAUGGGUCAAUUAAAAAUGAAGGCGG-UAUUAGUAUAAGAUGCAGAGCAGGCA .((((((((....))))))))(((((..(((.........)))..))))).....((...((.(-((((.......)))))...))...)) ( -22.50, z-score = -1.51, R) >droSec1.super_12 11949 90 - 2123299 AGAUAGAGAGUGCUCUCUAUCGAUUGCUCCAAAUUUAAGAUGGGUCAAUUAAAAAUGAAGGCGG-CAUUAGUAUAGGAUAUAGAGCAAGCA .......(..((((((.(((((((((..(((.........)))..)))))...((((.......-)))).......)))).))))))..). ( -20.40, z-score = -0.97, R) >droSim1.chr3R 18078434 90 - 27517382 AGAUAGAGAGUGCUCUCUAUCGAUUGCUCCAAAUUUAAGAUGGGUCAAUUAAAAAUGAAGGCGG-CAUUAGUAUAGGAUAUAGAGCAAGCA .......(..((((((.(((((((((..(((.........)))..)))))...((((.......-)))).......)))).))))))..). ( -20.40, z-score = -0.97, R) >consensus AGAUAGAGAGUGCUCUCUAUCGAUUGCUCCAAAUUUAAGAUGGGUCAAUUAAAAAUGAAGGCGG_CAUUAGUAUAGGAUACAGAGCAAGCA .......(..((((((.(((((((((.((((.........)))).)))))...((((........)))).......)))).))))))..). (-11.54 = -12.30 + 0.76)
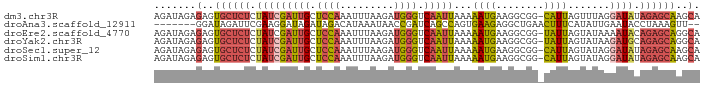
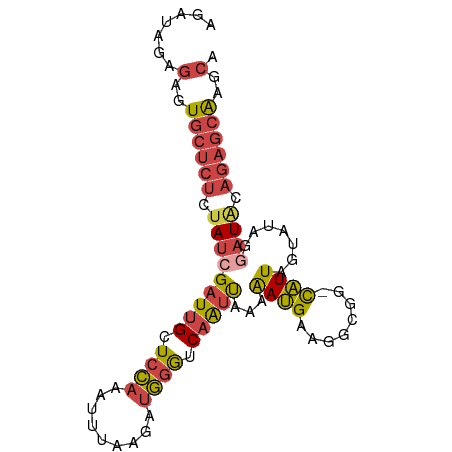
| Location | 12,037,277 – 12,037,367 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 76.83 |
| Shannon entropy | 0.44302 |
| G+C content | 0.37579 |
| Mean single sequence MFE | -18.91 |
| Consensus MFE | -11.26 |
| Energy contribution | -12.29 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.996462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
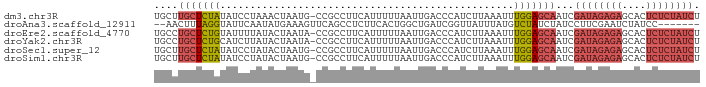
>dm3.chr3R 12037277 90 - 27905053 UGCUUGCUCUAUAUCCUAAACUAAUG-CCGCCUUCAUUUUUAAUUGACCCAUCUUAAAUUUGGAGCAAUCGAUAGAGAGCACUCUCUAUCU .(.((((((((.........(....)-......(((........))).............)))))))).)((((((((....)))))))). ( -18.70, z-score = -2.62, R) >droAna3.scaffold_12911 2858539 82 + 5364042 --AACUUUAGGUAUUCAAUAUGAAAGUUCAGCCUCUUCACUGGCUGAUCGGUUAUUUAUGUCUAUCUAUCCUUCGAAUCUAUCC------- --.....((((((..((..((((..(.((((((........)))))).)..))))...))..))))))................------- ( -18.50, z-score = -2.92, R) >droEre2.scaffold_4770 11560924 90 - 17746568 UGCCUGCUCUGUAUUUUAUACUAAUA-CCGCCUUCAUUUUUAAUUGACCCAUCUUAAAUUUGGAGCAAUCGAUAGAGAGCACUCUCUAUCU ....(((((((((((.......))))-).....(((........)))..............))))))...((((((((....)))))))). ( -20.60, z-score = -3.64, R) >droYak2.chr3R 14438071 90 + 28832112 UGCCUGCUCUGCAUCUUAUACUAAUA-CCGCCUUCAUUUUUAAUUGACCCAUCUUAAAUUUGGAGCAAUCGAUAGAGAGCACUCUCUAUCU ....((((((((..............-..))..(((........)))..............))))))...((((((((....)))))))). ( -18.09, z-score = -2.41, R) >droSec1.super_12 11949 90 + 2123299 UGCUUGCUCUAUAUCCUAUACUAAUG-CCGCCUUCAUUUUUAAUUGACCCAUCUUAAAUUUGGAGCAAUCGAUAGAGAGCACUCUCUAUCU .(.((((((((.......((..((((-.......))))..)).((((......))))...)))))))).)((((((((....)))))))). ( -18.80, z-score = -2.58, R) >droSim1.chr3R 18078434 90 + 27517382 UGCUUGCUCUAUAUCCUAUACUAAUG-CCGCCUUCAUUUUUAAUUGACCCAUCUUAAAUUUGGAGCAAUCGAUAGAGAGCACUCUCUAUCU .(.((((((((.......((..((((-.......))))..)).((((......))))...)))))))).)((((((((....)))))))). ( -18.80, z-score = -2.58, R) >consensus UGCUUGCUCUAUAUCCUAUACUAAUG_CCGCCUUCAUUUUUAAUUGACCCAUCUUAAAUUUGGAGCAAUCGAUAGAGAGCACUCUCUAUCU ....(((((((.................................................)))))))...((((((((....)))))))). (-11.26 = -12.29 + 1.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:19:14 2011