| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,031,903 – 12,031,954 |
| Length | 51 |
| Max. P | 0.852168 |

| Location | 12,031,903 – 12,031,954 |
|---|---|
| Length | 51 |
| Sequences | 9 |
| Columns | 56 |
| Reading direction | forward |
| Mean pairwise identity | 68.58 |
| Shannon entropy | 0.63390 |
| G+C content | 0.52823 |
| Mean single sequence MFE | -17.43 |
| Consensus MFE | -5.12 |
| Energy contribution | -5.14 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.67 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.852168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
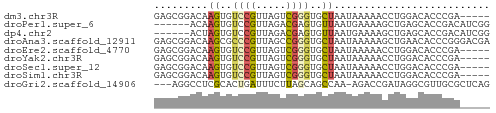
>dm3.chr3R 12031903 51 + 27905053 GAGCGGACAAGUGUCCGUUAGUCGGGUGCUAAUAAAAACCUGGACACCCGA----- .(((((((....)))))))..((((((((((.........))).)))))))----- ( -22.40, z-score = -4.07, R) >droPer1.super_6 606318 50 + 6141320 ------ACAAGUGUCCGUUAGACGAGUGUUAAUGAAAAGCUGAGCACCGACAUCGG ------....(((((((.....)).(((((..(....)....))))).)))))... ( -8.10, z-score = 0.30, R) >dp4.chr2 25278400 50 + 30794189 ------ACUAGUGUCCGUUAGACGAGUGUUAAUGAAAAGCUGAGCACCGACAUCGG ------.(..(((((((.....)).(((((..(....)....))))).)))))..) ( -9.80, z-score = -0.33, R) >droAna3.scaffold_12911 2853468 56 - 5364042 GAGCGGACAAGCGCCCGUUAGCCGGGUGCUAAUAAAAAGCUGAACACCCGGGACGA .(((((........)))))..(((((((................)))))))..... ( -17.49, z-score = -0.99, R) >droEre2.scaffold_4770 11555488 51 + 17746568 GAGCGGACAAGUGUCCGUUAGUCGGGUGCUAAUAAAAACCUGGACACCCGA----- .(((((((....)))))))..((((((((((.........))).)))))))----- ( -22.40, z-score = -4.07, R) >droYak2.chr3R 14432523 51 - 28832112 GAGCGGACAAGUGUCCGUUAGUCGGGUGCUAAUAAAAACCUGGACACCCGA----- .(((((((....)))))))..((((((((((.........))).)))))))----- ( -22.40, z-score = -4.07, R) >droSec1.super_12 6647 51 - 2123299 GAGCGGACAAGUGUCCGUUAGUCGGGUGCUAAUAAAAACCUGGACACCCGA----- .(((((((....)))))))..((((((((((.........))).)))))))----- ( -22.40, z-score = -4.07, R) >droSim1.chr3R 18073112 51 - 27517382 GAGCGGACAAGUGUCCGUUAGUCGGGUGCUAAUAAAAACCUGGACACCCGA----- .(((((((....)))))))..((((((((((.........))).)))))))----- ( -22.40, z-score = -4.07, R) >droGri2.scaffold_14906 13627941 52 - 14172833 ---AGGCCUCGCACUGAUUUCUUAGCAGCCAA-AGACCGAUAGGCGUUGCGCUCAG ---..((..(((.(((...((((........)-)))....))))))..))...... ( -9.50, z-score = 1.32, R) >consensus GAGCGGACAAGUGUCCGUUAGUCGGGUGCUAAUAAAAACCUGGACACCCGA_____ .........((..((((.....))))..)).......................... ( -5.12 = -5.14 + 0.03)

| Location | 12,031,903 – 12,031,954 |
|---|---|
| Length | 51 |
| Sequences | 9 |
| Columns | 56 |
| Reading direction | reverse |
| Mean pairwise identity | 68.58 |
| Shannon entropy | 0.63390 |
| G+C content | 0.52823 |
| Mean single sequence MFE | -15.56 |
| Consensus MFE | -5.49 |
| Energy contribution | -5.25 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 2.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.797048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
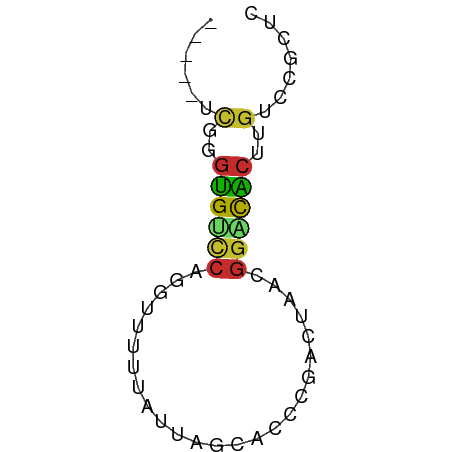
>dm3.chr3R 12031903 51 - 27905053 -----UCGGGUGUCCAGGUUUUUAUUAGCACCCGACUAACGGACACUUGUCCGCUC -----((((((((..............))))))))....(((((....)))))... ( -18.94, z-score = -3.21, R) >droPer1.super_6 606318 50 - 6141320 CCGAUGUCGGUGCUCAGCUUUUCAUUAACACUCGUCUAACGGACACUUGU------ ..(((((..(((..........)))..))).))(((.....)))......------ ( -6.80, z-score = 0.59, R) >dp4.chr2 25278400 50 - 30794189 CCGAUGUCGGUGCUCAGCUUUUCAUUAACACUCGUCUAACGGACACUAGU------ ..(((((..(((..........)))..))).))(((.....)))......------ ( -6.80, z-score = 0.57, R) >droAna3.scaffold_12911 2853468 56 + 5364042 UCGUCCCGGGUGUUCAGCUUUUUAUUAGCACCCGGCUAACGGGCGCUUGUCCGCUC .....(((((((((.(........).)))))))))....(((((....)))))... ( -20.30, z-score = -2.16, R) >droEre2.scaffold_4770 11555488 51 - 17746568 -----UCGGGUGUCCAGGUUUUUAUUAGCACCCGACUAACGGACACUUGUCCGCUC -----((((((((..............))))))))....(((((....)))))... ( -18.94, z-score = -3.21, R) >droYak2.chr3R 14432523 51 + 28832112 -----UCGGGUGUCCAGGUUUUUAUUAGCACCCGACUAACGGACACUUGUCCGCUC -----((((((((..............))))))))....(((((....)))))... ( -18.94, z-score = -3.21, R) >droSec1.super_12 6647 51 + 2123299 -----UCGGGUGUCCAGGUUUUUAUUAGCACCCGACUAACGGACACUUGUCCGCUC -----((((((((..............))))))))....(((((....)))))... ( -18.94, z-score = -3.21, R) >droSim1.chr3R 18073112 51 + 27517382 -----UCGGGUGUCCAGGUUUUUAUUAGCACCCGACUAACGGACACUUGUCCGCUC -----((((((((..............))))))))....(((((....)))))... ( -18.94, z-score = -3.21, R) >droGri2.scaffold_14906 13627941 52 + 14172833 CUGAGCGCAACGCCUAUCGGUCU-UUGGCUGCUAAGAAAUCAGUGCGAGGCCU--- ....(((...))).....(((((-(..((.(((.(....).))))))))))).--- ( -11.40, z-score = 1.16, R) >consensus _____UCGGGUGUCCAGGUUUUUAUUAGCACCCGACUAACGGACACUUGUCCGCUC ......(..((((((.........................))))))..)....... ( -5.49 = -5.25 + -0.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:19:11 2011