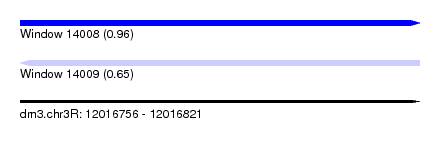
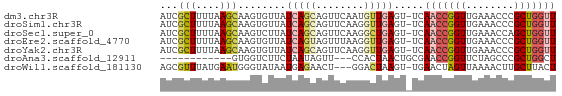
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,016,756 – 12,016,821 |
| Length | 65 |
| Max. P | 0.960268 |

| Location | 12,016,756 – 12,016,821 |
|---|---|
| Length | 65 |
| Sequences | 7 |
| Columns | 66 |
| Reading direction | forward |
| Mean pairwise identity | 71.27 |
| Shannon entropy | 0.58369 |
| G+C content | 0.43611 |
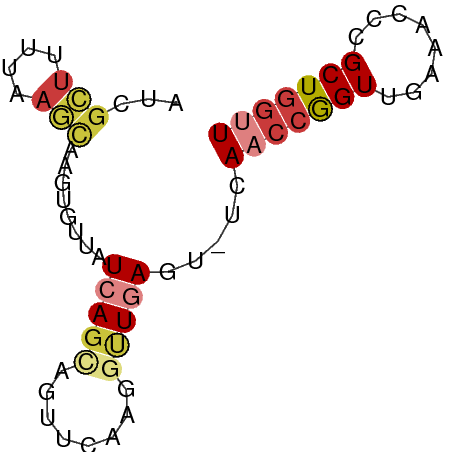
| Mean single sequence MFE | -15.03 |
| Consensus MFE | -5.88 |
| Energy contribution | -6.54 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12016756 65 + 27905053 AACCAGCGGGUUUCAACCGGUUGA-ACUCAACAUUGAACUGCUGAUAACACUUGCUUAAAAGCGAU ...((((((...((((...((((.-...)))).)))).)))))).......(((((....))))). ( -18.10, z-score = -3.11, R) >droSim1.chr3R 18057933 65 - 27517382 AACCAGCGGGUUUCAACCGGUUGA-ACUCAACCUUGAACUGCUGAUAACACUUGCUUAAAAGCGAU ...((((((...((((..(((((.-...))))))))).)))))).......(((((....))))). ( -20.40, z-score = -3.94, R) >droSec1.super_0 11177778 65 - 21120651 AACCAGCUGGUUUCAACCGGUUGA-ACUCAGCCUUGAACUGCUGAUAAGACUUGCUUAAAAGCGAU ...((((((((....)))))))).-..(((((........)))))......(((((....))))). ( -18.80, z-score = -2.74, R) >droEre2.scaffold_4770 11540452 65 + 17746568 AACCAGCGGGUUUCAACCGGUUGA-ACUCAACCUUAAACUACUGAUAACACUUGCUUAAAAGCGAU ...(((..(((....)))(((((.-...)))))........))).......(((((....))))). ( -13.40, z-score = -1.94, R) >droYak2.chr3R 14417164 65 - 28832112 AACCAGCGGGUUUCAACCGGUUGA-ACUCAACCUUGAACUGCUGAUAACACUUGCUUAAAAGCGAU ...((((((...((((..(((((.-...))))))))).)))))).......(((((....))))). ( -20.40, z-score = -3.94, R) >droAna3.scaffold_12911 2838700 51 - 5364042 AGCCAGCGGGCUAGAACCGGUUCGCAGUUAGUGG---AACUAUUAGAAGACCAC------------ .....(((((((......))))))).....((((---..((......)).))))------------ ( -11.80, z-score = -0.48, R) >droWil1.scaffold_181130 8840597 62 - 16660200 AGUAAGCAAGUUUUAACUAGUUCA-ACUUAGUCC---AGUUCUCAUUAUACCCAUUCAUAAACGCU ....(((........(((((....-..)))))..---........((((........))))..))) ( -2.30, z-score = 0.43, R) >consensus AACCAGCGGGUUUCAACCGGUUGA_ACUCAACCUUGAACUGCUGAUAACACUUGCUUAAAAGCGAU ...((((.(((....))).))))............................(((((....))))). ( -5.88 = -6.54 + 0.67)
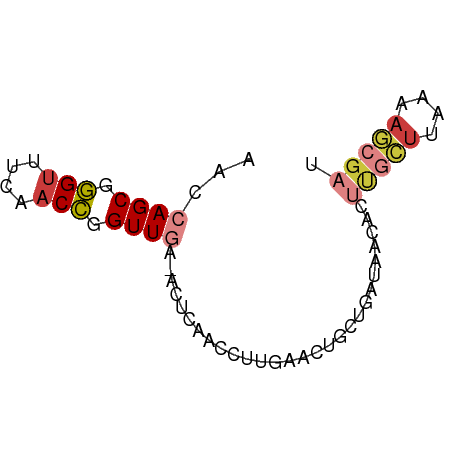
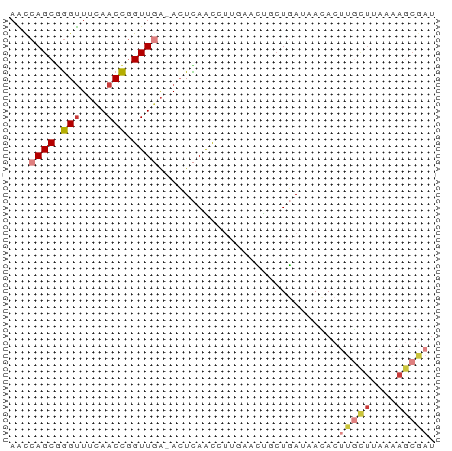
| Location | 12,016,756 – 12,016,821 |
|---|---|
| Length | 65 |
| Sequences | 7 |
| Columns | 66 |
| Reading direction | reverse |
| Mean pairwise identity | 71.27 |
| Shannon entropy | 0.58369 |
| G+C content | 0.43611 |
| Mean single sequence MFE | -15.07 |
| Consensus MFE | -6.28 |
| Energy contribution | -7.11 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.648846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12016756 65 - 27905053 AUCGCUUUUAAGCAAGUGUUAUCAGCAGUUCAAUGUUGAGU-UCAACCGGUUGAAACCCGCUGGUU ..(((((......)))))..((((((..((((((((((...-.))))..))))))....)))))). ( -16.10, z-score = -1.66, R) >droSim1.chr3R 18057933 65 + 27517382 AUCGCUUUUAAGCAAGUGUUAUCAGCAGUUCAAGGUUGAGU-UCAACCGGUUGAAACCCGCUGGUU ..(((((......)))))..((((((.......(((((...-.)))))(((....))).)))))). ( -18.20, z-score = -2.17, R) >droSec1.super_0 11177778 65 + 21120651 AUCGCUUUUAAGCAAGUCUUAUCAGCAGUUCAAGGCUGAGU-UCAACCGGUUGAAACCAGCUGGUU ...(((....)))........(((((........)))))..-..(((((((((....))))))))) ( -19.40, z-score = -2.41, R) >droEre2.scaffold_4770 11540452 65 - 17746568 AUCGCUUUUAAGCAAGUGUUAUCAGUAGUUUAAGGUUGAGU-UCAACCGGUUGAAACCCGCUGGUU ..(((((......)))))..((((((.((((..(((((...-.))))).....))))..)))))). ( -16.00, z-score = -1.76, R) >droYak2.chr3R 14417164 65 + 28832112 AUCGCUUUUAAGCAAGUGUUAUCAGCAGUUCAAGGUUGAGU-UCAACCGGUUGAAACCCGCUGGUU ..(((((......)))))..((((((.......(((((...-.)))))(((....))).)))))). ( -18.20, z-score = -2.17, R) >droAna3.scaffold_12911 2838700 51 + 5364042 ------------GUGGUCUUCUAAUAGUU---CCACUAACUGCGAACCGGUUCUAGCCCGCUGGCU ------------((((.((......))..---)))).(((((.....)))))..((((....)))) ( -10.30, z-score = 0.01, R) >droWil1.scaffold_181130 8840597 62 + 16660200 AGCGUUUAUGAAUGGGUAUAAUGAGAACU---GGACUAAGU-UGAACUAGUUAAAACUUGCUUACU ............((((((.......((((---((.......-....))))))......)))))).. ( -7.32, z-score = 0.36, R) >consensus AUCGCUUUUAAGCAAGUGUUAUCAGCAGUUCAAGGUUGAGU_UCAACCGGUUGAAACCCGCUGGUU ...(((....)))........(((((........))))).....(((((((........))))))) ( -6.28 = -7.11 + 0.83)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:19:09 2011