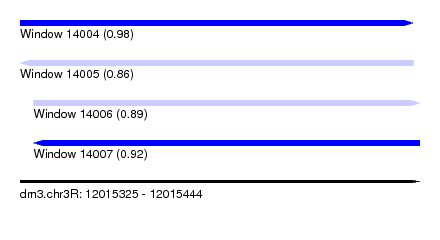
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,015,325 – 12,015,444 |
| Length | 119 |
| Max. P | 0.983068 |

| Location | 12,015,325 – 12,015,442 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 50.99 |
| Shannon entropy | 0.69134 |
| G+C content | 0.48947 |
| Mean single sequence MFE | -24.99 |
| Consensus MFE | -14.23 |
| Energy contribution | -14.90 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.983068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
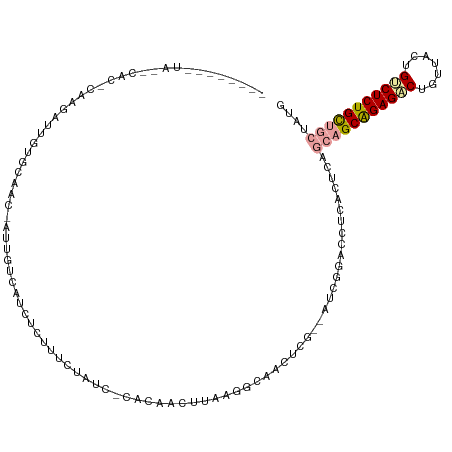
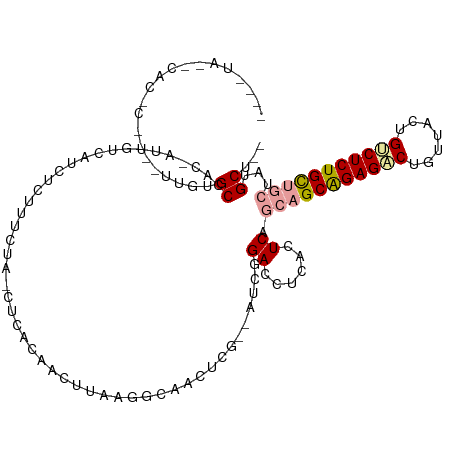
>dm3.chr3R 12015325 117 + 27905053 CUAAUGGCUAUACACACUGGCGUGUGCCAC-AUUGUCAUCUCUCUCUAACACACUAUUUAUGACCACUCG--AUCGGACCUCACUCAGCUGCAGAGGCUGUUCGUGCCUCUGCCGCAGCG ......(((.......((((((.(((....-...(((((....................)))))))).))--.))))..........((.((((((((.......)))))))).))))). ( -31.16, z-score = -1.06, R) >droAna3.scaffold_12966 14940 103 + 443544 ----------------CAAGUUGAGCCUCC-AUCGUCAUCUCUUUCCAUCUCACAACUUUCGGUAACUCUUCAACGGACCUCUCUCACCAGCAGAGACUGCUACCGUCUCUGCUGCUAUG ----------------...((((((.....-((((.........................)))).....)))))).............((((((((((.......))))))))))..... ( -22.61, z-score = -2.25, R) >droVir3.scaffold_13324 1398567 113 + 2960039 ----CAAUUAAUCACUCAAAAUUGUGCAACUAUUGUUCGCCGUUGCUGUU-CUUCAGUCAAAGCUGUACG--AUCAGACCUCACUCAGCAGCGGAGUCUCUCUUUGUCUCUGUUACUAUG ----(((......((((......(((.(((....))))))((((((((..-....(((.....(((....--..))).....)))))))))))))))......))).............. ( -21.20, z-score = -0.19, R) >consensus ________UA__CAC_CAAGAUUGUGCAAC_AUUGUCAUCUCUUUCUAUC_CACAACUUAAGGCAACUCG__AUCGGACCUCACUCAGCAGCAGAGACUGUUACUGUCUCUGCUGCUAUG .......................................................................................(((((((((((.......))))))))))).... (-14.23 = -14.90 + 0.67)

| Location | 12,015,325 – 12,015,442 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 50.99 |
| Shannon entropy | 0.69134 |
| G+C content | 0.48947 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -13.84 |
| Energy contribution | -14.31 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12015325 117 - 27905053 CGCUGCGGCAGAGGCACGAACAGCCUCUGCAGCUGAGUGAGGUCCGAU--CGAGUGGUCAUAAAUAGUGUGUUAGAGAGAGAUGACAAU-GUGGCACACGCCAGUGUGUAUAGCCAUUAG ((((((.((((((((.......)))))))).))..)))).........--..((((((((((....((((((((.(..(......)..)-.)))))))).....))))....)))))).. ( -38.60, z-score = -1.21, R) >droAna3.scaffold_12966 14940 103 - 443544 CAUAGCAGCAGAGACGGUAGCAGUCUCUGCUGGUGAGAGAGGUCCGUUGAAGAGUUACCGAAAGUUGUGAGAUGGAAAGAGAUGACGAU-GGAGGCUCAACUUG---------------- ..((.((((((((((.......)))))))))).)).(((...(((((((.....(((((....)..))))..(....).......))))-)))..)))......---------------- ( -32.20, z-score = -2.30, R) >droVir3.scaffold_13324 1398567 113 - 2960039 CAUAGUAACAGAGACAAAGAGAGACUCCGCUGCUGAGUGAGGUCUGAU--CGUACAGCUUUGACUGAAG-AACAGCAACGGCGAACAAUAGUUGCACAAUUUUGAGUGAUUAAUUG---- ........(((((.....((.((((..((((....))))..))))..)--)......)))))(((.(((-(...(((((...........)))))....)))).))).........---- ( -25.20, z-score = -0.04, R) >consensus CAUAGCAGCAGAGACAAAAACAGACUCUGCUGCUGAGUGAGGUCCGAU__CGAGUAGCCAUAAAUAGUG_GAUAGAAAGAGAUGACAAU_GUAGCACAAACUUG_GUG__UA________ ..(((((((((((((.......))))))))))))).......((((((......((....))........))))))............................................ (-13.84 = -14.31 + 0.46)

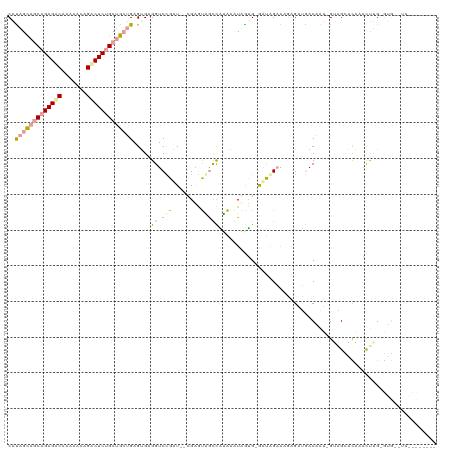
| Location | 12,015,329 – 12,015,444 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 53.41 |
| Shannon entropy | 0.65517 |
| G+C content | 0.49191 |
| Mean single sequence MFE | -24.35 |
| Consensus MFE | -14.46 |
| Energy contribution | -15.13 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.70 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.887753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
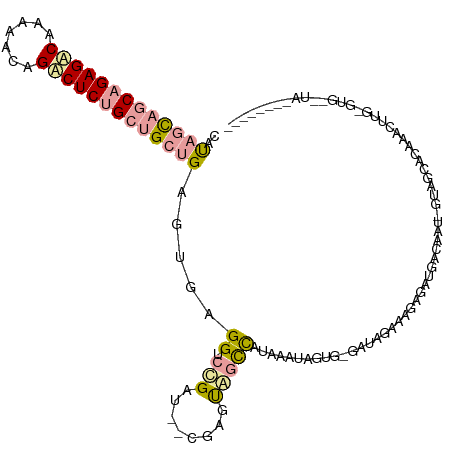
>dm3.chr3R 12015329 115 + 27905053 UGGCUAUACACACUGGCGUGUGCCAC-AUUGUCAUCUCUCUCUAACACACUAUUUAUGACCACUCG--AUCGGACCUCACUCAGCUGCAGAGGCUGUUCGUGCCUCUGCCGCAGCGCU-- ..(((.......((((((.(((....-...(((((....................)))))))).))--.))))..........((.((((((((.......)))))))).)))))...-- ( -31.16, z-score = -0.52, R) >droAna3.scaffold_12966 14944 103 + 443544 -----------------UUGAGCCUCCAUCGUCAUCUCUUUCCAUCUCACAACUUUCGGUAACUCUUCAACGGACCUCUCUCACCAGCAGAGACUGCUACCGUCUCUGCUGCUAUGCUUU -----------------..((((...............................((((............))))..........((((((((((.......))))))))))....)))). ( -21.00, z-score = -1.91, R) >droVir3.scaffold_13324 1398571 111 + 2960039 ----UAAUCACUCAAAAUUGUGCAACUAUUGUUCGCCGUUGCUG-UUCUUCAGUCAAAGCUGUACG--AUCAGACCUCACUCAGCAGCGGAGUCUCUCUUUGUCUCUGUUACUAUGCU-- ----.....((((......(((.(((....))))))((((((((-......(((.....(((....--..))).....))))))))))))))).........................-- ( -20.90, z-score = 0.34, R) >consensus ____UA__CAC_C____UUGUGCCAC_AUUGUCAUCUCUUUCUA_CUCACAACUUAAGGCAACUCG__AUCGGACCUCACUCAGCAGCAGAGACUGUUACUGUCUCUGCUGCUAUGCU__ .....................((.................................................((......)).(((((((((((.......)))))))))))...))... (-14.46 = -15.13 + 0.67)

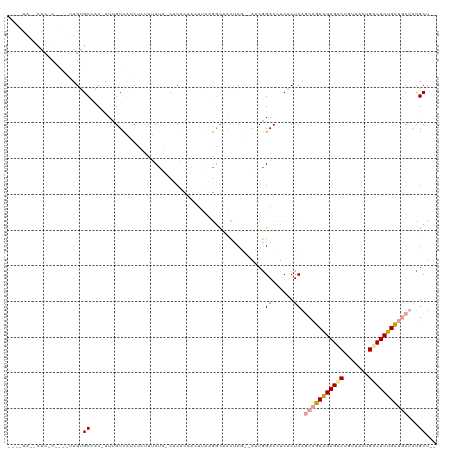
| Location | 12,015,329 – 12,015,444 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 53.41 |
| Shannon entropy | 0.65517 |
| G+C content | 0.49191 |
| Mean single sequence MFE | -32.17 |
| Consensus MFE | -16.08 |
| Energy contribution | -16.54 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.918195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12015329 115 - 27905053 --AGCGCUGCGGCAGAGGCACGAACAGCCUCUGCAGCUGAGUGAGGUCCGAU--CGAGUGGUCAUAAAUAGUGUGUUAGAGAGAGAUGACAAU-GUGGCACACGCCAGUGUGUAUAGCCA --.(((((((.((((((((.......)))))))).)).......(((..(((--(....)))).......((((((((.(..(......)..)-.))))))))))))))))......... ( -39.10, z-score = -0.98, R) >droAna3.scaffold_12966 14944 103 - 443544 AAAGCAUAGCAGCAGAGACGGUAGCAGUCUCUGCUGGUGAGAGAGGUCCGUUGAAGAGUUACCGAAAGUUGUGAGAUGGAAAGAGAUGACGAUGGAGGCUCAA----------------- ......((.((((((((((.......)))))))))).)).(((...(((((((.....(((((....)..))))..(....).......)))))))..)))..----------------- ( -32.20, z-score = -2.74, R) >droVir3.scaffold_13324 1398571 111 - 2960039 --AGCAUAGUAACAGAGACAAAGAGAGACUCCGCUGCUGAGUGAGGUCUGAU--CGUACAGCUUUGACUGAAGAA-CAGCAACGGCGAACAAUAGUUGCACAAUUUUGAGUGAUUA---- --..........(((((.....((.((((..((((....))))..))))..)--)......)))))(((.((((.-..(((((...........)))))....)))).))).....---- ( -25.20, z-score = 0.27, R) >consensus __AGCAUAGCAGCAGAGACAAAAACAGACUCUGCUGCUGAGUGAGGUCCGAU__CGAGUAGCCAUAAAUAGUGAG_UAGAAAGAGAUGACAAU_GUGGCACAA____G_GUG__UA____ ...((.(((((((((((((.......))))))))))))).......((((((......((....))........)))))).................))..................... (-16.08 = -16.54 + 0.46)

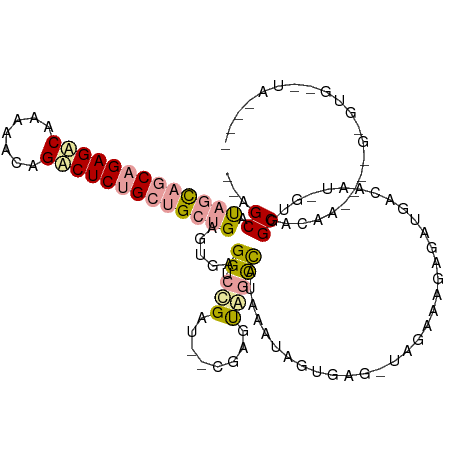
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:19:08 2011