
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,008,838 – 12,008,939 |
| Length | 101 |
| Max. P | 0.771634 |

| Location | 12,008,838 – 12,008,939 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 73.19 |
| Shannon entropy | 0.50589 |
| G+C content | 0.39165 |
| Mean single sequence MFE | -20.57 |
| Consensus MFE | -8.41 |
| Energy contribution | -9.02 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.771634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
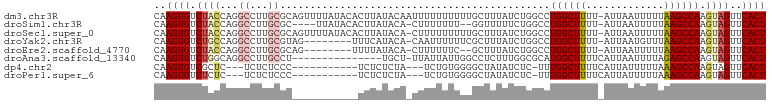
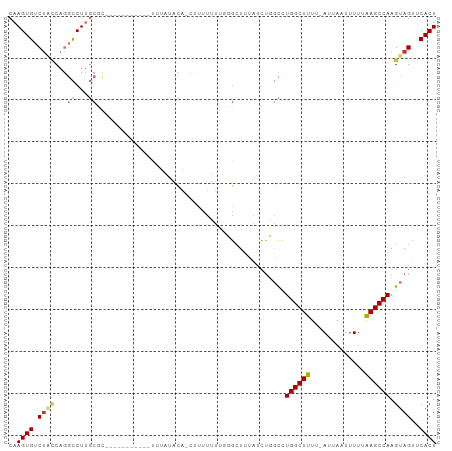
>dm3.chr3R 12008838 101 + 27905053 CAAGUGUCUACCAGGCCUUGCGCAGUUUUAUACACUUAUACAAUUUUUUUUUUGCUUUAUCUGGCCUGGCUUUU-AUUAAUUUUUAAGCCAAGUAGUUCACU ..((((.((((.(((((....((((..........................)))).......)))))(((((..-(......)..)))))..))))..)))) ( -21.87, z-score = -1.99, R) >droSim1.chr3R 18050476 94 - 27517382 CAAGUGUCUACCAGGCCUUGCGC----UUAUACACUUAUACA-CUUUUUUU--GGUUUUUCUGGCCUGGCUUUU-AUUAAUUUUUAAGCCAAGUAGUUCACU ..((((.((((.(((((......----.((((....)))).(-((......--)))......)))))(((((..-(......)..)))))..))))..)))) ( -20.30, z-score = -1.53, R) >droSec1.super_0 11170315 100 - 21120651 CAAGUGUCUACCAGGCCUUGCGCAGUUUUAUACACUUAUACA-CUUUUUUUUUGCUUUAUCUGGCCUGGCUUUU-AUUAAUUUUUAAGCCAAGUAGUUCACU ..((((.((((.(((((....((((.................-........)))).......)))))(((((..-(......)..)))))..))))..)))) ( -21.91, z-score = -2.00, R) >droYak2.chr3R 14409712 92 - 28832112 CAAGUGUCUGCCAGGCCUUGCGUAG--------UUUCAUACA-CAAUUUUUUCGCUUUAUCUGGCCUGGCUUUU-AUUAAUGUUUAAGCCAAGUAGUUCACU ..((((.((((.(((((..(((.((--------((.......-.))))....))).......)))))(((((..-((....))..)))))..))))..)))) ( -23.60, z-score = -2.40, R) >droEre2.scaffold_4770 11533005 90 + 17746568 CAAGUGUCUACCAGGCCUUGCGCAG--------UUUUAUACA-CUUUUUUC--GCUUUAUCUGGCCUGGCUUUU-AUUAAUUUUUAAGCCAAGUAGUUCACU ..((((.((((.(((((..(((.((--------.........-....)).)--)).......)))))(((((..-(......)..)))))..))))..)))) ( -21.72, z-score = -2.36, R) >droAna3.scaffold_13340 2135980 86 - 23697760 CAAGUGUCUGGCAGGCCUUGCCU---------------UGCU-UUAUUAUUGGCCUCUUUGGCGCAUGGCUUUUCAUUAAUUUUAGAGCCAAGUAGUUCACU ...(((((.((.(((((......---------------....-........)))))))..))))).(((((((...........)))))))........... ( -23.07, z-score = -0.42, R) >dp4.chr2 4738105 84 - 30794189 CAAGUGUCGCUC---UCUCUCCC-----------UCUCUCUA---UCUGUGGGGCUAUAUCUC-UUUGGCUUUUCAUUAUUUUUAAAGCCAAGUAGUUCACU ..((((..(((.---....((((-----------.(......---...).)))).........-(((((((((...........))))))))).))).)))) ( -16.20, z-score = -2.37, R) >droPer1.super_6 4753475 84 - 6141320 CAAGUGUCUCUC---UCUCUCCC-----------UCUCUCUA---UCUGUGGGGCUAUAUCUC-UUUGGCUUUUCAUUAUUUUUAAAGCCAAGUAGUUCACU ..((((......---.....(((-----------.(......---...).)))(((((.....-.((((((((...........))))))))))))).)))) ( -15.90, z-score = -2.66, R) >consensus CAAGUGUCUACCAGGCCUUGCGC___________UUUAUACA_CUUUUUUUGGGCUUUAUCUGGCCUGGCUUUU_AUUAAUUUUUAAGCCAAGUAGUUCACU ..((((.((((...((...)).............................................((((((.............)))))).))))..)))) ( -8.41 = -9.02 + 0.61)

| Location | 12,008,838 – 12,008,939 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 73.19 |
| Shannon entropy | 0.50589 |
| G+C content | 0.39165 |
| Mean single sequence MFE | -21.47 |
| Consensus MFE | -7.67 |
| Energy contribution | -7.79 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.738620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
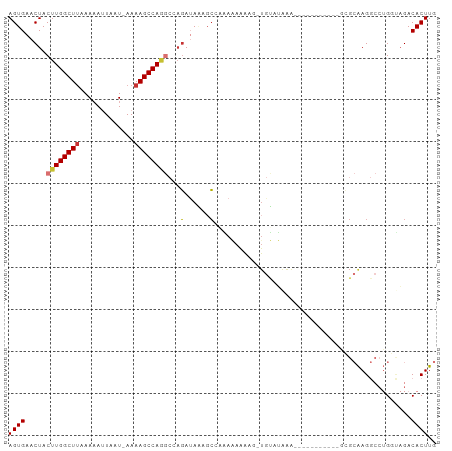
>dm3.chr3R 12008838 101 - 27905053 AGUGAACUACUUGGCUUAAAAAUUAAU-AAAAGCCAGGCCAGAUAAAGCAAAAAAAAAAUUGUAUAAGUGUAUAAAACUGCGCAAGGCCUGGUAGACACUUG ((((..(((((.(((((..........-..)))))(((((.......((((........))))....(((((......)))))..)))))))))).)))).. ( -26.50, z-score = -3.14, R) >droSim1.chr3R 18050476 94 + 27517382 AGUGAACUACUUGGCUUAAAAAUUAAU-AAAAGCCAGGCCAGAAAAACC--AAAAAAAG-UGUAUAAGUGUAUAA----GCGCAAGGCCUGGUAGACACUUG ((((..(((((.(((((..........-..)))))(((((......((.--.......)-)......((((....----))))..)))))))))).)))).. ( -22.90, z-score = -2.10, R) >droSec1.super_0 11170315 100 + 21120651 AGUGAACUACUUGGCUUAAAAAUUAAU-AAAAGCCAGGCCAGAUAAAGCAAAAAAAAAG-UGUAUAAGUGUAUAAAACUGCGCAAGGCCUGGUAGACACUUG ((((..(((((.(((((..........-..)))))(((((.......(((.........-)))....(((((......)))))..)))))))))).)))).. ( -24.80, z-score = -2.30, R) >droYak2.chr3R 14409712 92 + 28832112 AGUGAACUACUUGGCUUAAACAUUAAU-AAAAGCCAGGCCAGAUAAAGCGAAAAAAUUG-UGUAUGAAA--------CUACGCAAGGCCUGGCAGACACUUG ((((.......((.......)).....-....((((((((.(......).......(((-((((.....--------.)))))))))))))))...)))).. ( -25.70, z-score = -3.16, R) >droEre2.scaffold_4770 11533005 90 - 17746568 AGUGAACUACUUGGCUUAAAAAUUAAU-AAAAGCCAGGCCAGAUAAAGC--GAAAAAAG-UGUAUAAAA--------CUGCGCAAGGCCUGGUAGACACUUG ((((..(((((.(((((..........-..)))))(((((.(......)--.......(-((((.....--------.)))))..)))))))))).)))).. ( -23.60, z-score = -2.49, R) >droAna3.scaffold_13340 2135980 86 + 23697760 AGUGAACUACUUGGCUCUAAAAUUAAUGAAAAGCCAUGCGCCAAAGAGGCCAAUAAUAA-AGCA---------------AGGCAAGGCCUGCCAGACACUUG ((((.......(((((...............)))))((.(((.....))))).......-.(((---------------.(((...))))))....)))).. ( -17.66, z-score = 0.36, R) >dp4.chr2 4738105 84 + 30794189 AGUGAACUACUUGGCUUUAAAAAUAAUGAAAAGCCAAA-GAGAUAUAGCCCCACAGA---UAGAGAGA-----------GGGAGAGA---GAGCGACACUUG ((((..((..((((((((...........)))))))).-.)).......(((.....---........-----------))).....---......)))).. ( -15.32, z-score = -2.17, R) >droPer1.super_6 4753475 84 + 6141320 AGUGAACUACUUGGCUUUAAAAAUAAUGAAAAGCCAAA-GAGAUAUAGCCCCACAGA---UAGAGAGA-----------GGGAGAGA---GAGAGACACUUG ((((..((..((((((((...........)))))))).-.)).......(((.....---........-----------))).....---......)))).. ( -15.32, z-score = -2.55, R) >consensus AGUGAACUACUUGGCUUAAAAAUUAAU_AAAAGCCAGGCCAGAUAAAGCCAAAAAAAAG_UGUAUAAA___________GCGCAAGGCCUGGUAGACACUUG ((((.....((((((((.............))))))))...(......)...............................................)))).. ( -7.67 = -7.79 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:19:05 2011