| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,602,474 – 7,602,594 |
| Length | 120 |
| Max. P | 0.808059 |

| Location | 7,602,474 – 7,602,574 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 66.25 |
| Shannon entropy | 0.66141 |
| G+C content | 0.40943 |
| Mean single sequence MFE | -23.97 |
| Consensus MFE | -7.22 |
| Energy contribution | -7.03 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.556474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
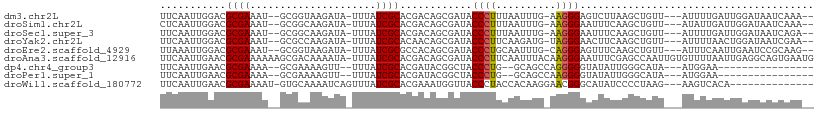
>dm3.chr2L 7602474 100 + 23011544 UUCAAUUGGACGCGAAAU--GCGGUAAGAUA-UUUAUCGCACGACAGCGAUACCCUUUAAUUUG-AAGGGAGUCUUAAGCUGUU---AUUUUGAUUGGAUAAUCAAA-- ((((((..((.......(--((((((((...-))))))))).(((((((((.((((((.....)-))))).)))....))))))---..))..))))))........-- ( -31.00, z-score = -4.00, R) >droSim1.chr2L 7399209 100 + 22036055 CUCAAUUGGACGCGAAAU--GCGGCAAGAUA-UUUAUCGCACGACAGCGAUACCCUUUAAUUUG-AAGGGAAUUUCAAGCUGUU---AUAUUGAUUGGAUAAUCAAA-- .(((((..(.(((.....--)))((......-..((((((......))))))((((((.....)-)))))........))....---...)..))))).........-- ( -24.20, z-score = -1.83, R) >droSec1.super_3 3116028 100 + 7220098 UUCAAUUGGACGCGAAAU--GCGGCAAGAUA-UUUAUCGCACGACAGCGAUACCCUUUAAUUUG-AAGGGAAUUUCAAGCUGUU---AUUUUGAUUGGAUAAUCAGA-- ((((((..((........--(((((......-..((((((......))))))((((((.....)-)))))........))))).---..))..))))))........-- ( -26.40, z-score = -2.46, R) >droYak2.chr2L 17031266 100 - 22324452 UUCAAUUGGACGCGAAAU--GCGCCAAGAUA-UUUAUCGCACAACAGCGAUACCCUUCAAGAUG-UAGGGAACUUCAAGCUGUU---AUUUUAACUGGAUAAUCGAA-- .....((((.(((.....--)))))))....-..((((((......))))))((((.(.....)-.)))).........(.(((---((((.....))))))).)..-- ( -22.00, z-score = -1.19, R) >droEre2.scaffold_4929 16521748 100 + 26641161 UUAAAUUGGACGCGAAAU--GCGGUAAGAUA-UUUAUCGCGCCACAGCGAUACCCUGCAAUUUG-CAGGGAGUUUCAAGCUGUU---AUUUCAAUUGAAUCCGCAAG-- .......(((..(((...--((((((((...-))))))))...((((((((.((((((.....)-))))).)))....))))).---.......)))..))).....-- ( -30.20, z-score = -2.59, R) >droAna3.scaffold_12916 15533023 108 + 16180835 UUCAAUUGAACGCGAAAAAAGCGACAAAAUA-UUUAUCGCACGACAGCGAUACCCUUCAAUUUACAAGGGAAUUUCGAGCCAAUUGUGUUUUAAUUGAGGCAGUGAAUG (((((((((((((((......(((.......-..((((((......))))))(((((........)))))....)))......))))).)))))))))).......... ( -27.20, z-score = -2.65, R) >dp4.chr4_group3 1490040 84 + 11692001 UUCAAUUGAACGCGAAAA--GCGAAAAGUU--UUUAUCGCACGAUACGGCUACCCUG--GCAGCCAGGGGGUAUAUUGGGCAUA---AUGGAA---------------- ((((.......(((((((--((.....)))--))..)))).(((((..(((.(((((--(...))))))))).)))))......---.)))).---------------- ( -23.60, z-score = -1.50, R) >droPer1.super_1 2958430 84 + 10282868 UUCAAUUGAACGCGAAAA--GCGAAAAGUU--UUUAUCGCACGAUACGGCUACCCUG--GCAGCCAAGGGGUAUAUUGGGCAUA---AUGGAA---------------- ((((..((..((((((((--((.....)))--))..)))).......((((.(....--).))))............)..))..---.)))).---------------- ( -18.20, z-score = 0.19, R) >droWil1.scaffold_180772 3425700 91 + 8906247 UUCAAUUGAACGCGAAAAU-GUGCAAAAUCAGUUUAUCGCACGAAAUGGUUACCCUACCACAAGGAACGGGCAUAUCCCCUAAG---AAGUCACA-------------- (((..(((...((((((((-...........)))).))))......((((......))))))).....(((......)))...)---))......-------------- ( -12.90, z-score = 1.00, R) >consensus UUCAAUUGGACGCGAAAU__GCGGCAAGAUA_UUUAUCGCACGACAGCGAUACCCUUCAAUUUG_AAGGGAAUUUCAAGCUGUU___AUUUUAAUUGGAUAAUCAAA__ ....................((((.(((....))).))))............((((..........))))....................................... ( -7.22 = -7.03 + -0.18)


| Location | 7,602,474 – 7,602,574 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 66.25 |
| Shannon entropy | 0.66141 |
| G+C content | 0.40943 |
| Mean single sequence MFE | -21.97 |
| Consensus MFE | -8.33 |
| Energy contribution | -8.12 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.808059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
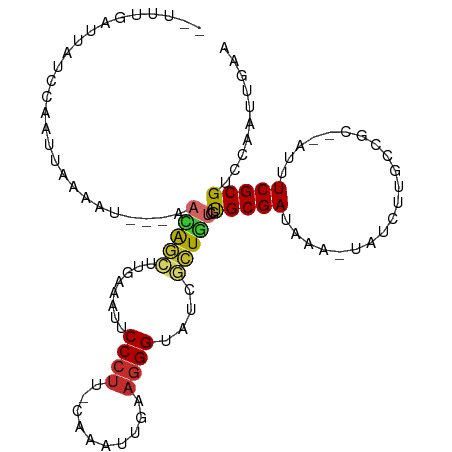
>dm3.chr2L 7602474 100 - 23011544 --UUUGAUUAUCCAAUCAAAAU---AACAGCUUAAGACUCCCUU-CAAAUUAAAGGGUAUCGCUGUCGUGCGAUAAA-UAUCUUACCGC--AUUUCGCGUCCAAUUGAA --(((((((....)))))))..---.......(((((..(((((-.......)))))((((((......))))))..-..))))).(((--.....))).......... ( -20.80, z-score = -2.38, R) >droSim1.chr2L 7399209 100 - 22036055 --UUUGAUUAUCCAAUCAAUAU---AACAGCUUGAAAUUCCCUU-CAAAUUAAAGGGUAUCGCUGUCGUGCGAUAAA-UAUCUUGCCGC--AUUUCGCGUCCAAUUGAG --.............(((((..---....((..((....(((((-.......)))))((((((......))))))..-..))..))(((--.....)))....))))). ( -21.10, z-score = -1.80, R) >droSec1.super_3 3116028 100 - 7220098 --UCUGAUUAUCCAAUCAAAAU---AACAGCUUGAAAUUCCCUU-CAAAUUAAAGGGUAUCGCUGUCGUGCGAUAAA-UAUCUUGCCGC--AUUUCGCGUCCAAUUGAA --.............((((...---....((..((....(((((-.......)))))((((((......))))))..-..))..))(((--.....))).....)))). ( -20.10, z-score = -1.65, R) >droYak2.chr2L 17031266 100 + 22324452 --UUCGAUUAUCCAGUUAAAAU---AACAGCUUGAAGUUCCCUA-CAUCUUGAAGGGUAUCGCUGUUGUGCGAUAAA-UAUCUUGGCGC--AUUUCGCGUCCAAUUGAA --.((((((.....(((...((---((((((..((....((((.-(.....).))))..))))))))))..)))...-......(((((--.....))))).)))))). ( -24.10, z-score = -1.44, R) >droEre2.scaffold_4929 16521748 100 - 26641161 --CUUGCGGAUUCAAUUGAAAU---AACAGCUUGAAACUCCCUG-CAAAUUGCAGGGUAUCGCUGUGGCGCGAUAAA-UAUCUUACCGC--AUUUCGCGUCCAAUUUAA --..(((((.((((((((....---..))).)))))...(((((-(.....))))))((((((......))))))..-.......))))--)................. ( -29.20, z-score = -2.54, R) >droAna3.scaffold_12916 15533023 108 - 16180835 CAUUCACUGCCUCAAUUAAAACACAAUUGGCUCGAAAUUCCCUUGUAAAUUGAAGGGUAUCGCUGUCGUGCGAUAAA-UAUUUUGUCGCUUUUUUCGCGUUCAAUUGAA .......................((((((((.(((((..(((((........)))))((((((......))))))..-..............))))).).))))))).. ( -24.30, z-score = -1.72, R) >dp4.chr4_group3 1490040 84 - 11692001 ----------------UUCCAU---UAUGCCCAAUAUACCCCCUGGCUGC--CAGGGUAGCCGUAUCGUGCGAUAAA--AACUUUUCGC--UUUUCGCGUUCAAUUGAA ----------------.((.((---(((((..............((((((--....)))))).......((((....--......))))--.....))))..))).)). ( -18.00, z-score = -0.72, R) >droPer1.super_1 2958430 84 - 10282868 ----------------UUCCAU---UAUGCCCAAUAUACCCCUUGGCUGC--CAGGGUAGCCGUAUCGUGCGAUAAA--AACUUUUCGC--UUUUCGCGUUCAAUUGAA ----------------.((.((---(((((..............((((((--....)))))).......((((....--......))))--.....))))..))).)). ( -18.00, z-score = -0.67, R) >droWil1.scaffold_180772 3425700 91 - 8906247 --------------UGUGACUU---CUUAGGGGAUAUGCCCGUUCCUUGUGGUAGGGUAACCAUUUCGUGCGAUAAACUGAUUUUGCAC-AUUUUCGCGUUCAAUUGAA --------------.((((...---.....(..((.(((((...((....))..)))))...))..)((((((..........))))))-....))))........... ( -22.10, z-score = -0.48, R) >consensus __UUUGAUUAUCCAAUUAAAAU___AACAGCUUGAAAUUCCCUU_CAAAUUGAAGGGUAUCGCUGUCGUGCGAUAAA_UAUCUUGCCGC__AUUUCGCGUCCAAUUGAA ..........................(((((........((((..........))))....)))))..(((((.....................))))).......... ( -8.33 = -8.12 + -0.21)


| Location | 7,602,494 – 7,602,594 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 56.58 |
| Shannon entropy | 0.86713 |
| G+C content | 0.41049 |
| Mean single sequence MFE | -20.24 |
| Consensus MFE | -4.37 |
| Energy contribution | -4.26 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.508830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
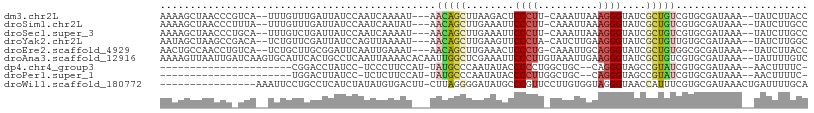
>dm3.chr2L 7602494 100 - 23011544 AAAAGCUAACCCGUCA--UUUGUUUGAUUAUCCAAUCAAAAU---AACAGCUUAAGACUCCCUU-CAAAUUAAAGGGUAUCGCUGUCGUGCGAUAAA--UAUCUUACC ..(((((....((...--..))(((((((....)))))))..---...)))))((((..(((((-.......)))))((((((......))))))..--..))))... ( -20.00, z-score = -2.94, R) >droSim1.chr2L 7399229 100 - 22036055 AAAAGCUAACCCUUUA--UUUGUUUGAUUAUCCAAUCAAUAU---AACAGCUUGAAAUUCCCUU-CAAAUUAAAGGGUAUCGCUGUCGUGCGAUAAA--UAUCUUGCC ....((..((((((((--.((((((((((....)))))).))---))....(((((......))-)))..))))))))(((((......)))))...--......)). ( -22.00, z-score = -3.38, R) >droSec1.super_3 3116048 100 - 7220098 AAAAGCUAACCCUGCA--UUUGUCUGAUUAUCCAAUCAAAAU---AACAGCUUGAAAUUCCCUU-CAAAUUAAAGGGUAUCGCUGUCGUGCGAUAAA--UAUCUUGCC ....((..(((((((.--.((((.(((((....)))))..))---))..))(((((......))-))).....)))))(((((......)))))...--......)). ( -19.40, z-score = -2.22, R) >droYak2.chr2L 17031286 100 + 22324452 AAUAGCUAAGCCGACA--UCUGUUCGAUUAUCCAGUUAAAAU---AACAGCUUGAAGUUCCCUA-CAUCUUGAAGGGUAUCGCUGUUGUGCGAUAAA--UAUCUUGGC ....((((((.((((.--...).))).(((((..((....((---((((((..((....((((.-(.....).))))..))))))))))))))))).--...)))))) ( -21.30, z-score = -0.65, R) >droEre2.scaffold_4929 16521768 100 - 26641161 AACUGCCAACCUGUCA--UCUGCUUGCGGAUUCAAUUGAAAU---AACAGCUUGAAACUCCCUG-CAAAUUGCAGGGUAUCGCUGUGGCGCGAUAAA--UAUCUUACC ....(((....((..(--((((....))))).))........---.(((((..((....(((((-(.....))))))..))))))))))........--......... ( -26.60, z-score = -2.14, R) >droAna3.scaffold_12916 15533045 106 - 16180835 AAAAGUUAAUUGAUCAAGUGCAUUCACUGCCUCAAUUAAAACACAAUUGGCUCGAAAUUCCCUUGUAAAUUGAAGGGUAUCGCUGUCGUGCGAUAAA--UAUUUUGUC .....((((((((...((((....))))...)))))))).............((((((.(((((........)))))((((((......))))))..--.)))))).. ( -23.70, z-score = -1.94, R) >dp4.chr4_group3 1490060 79 - 11692001 ----------------------CGGACCUAUCC-UCCCUUCCAU-UAUGCCCAAUAUACCCCCUGGCUGC--CAGGGUAGCCGUAUCGUGCGAUAAA--AACUUUUC- ----------------------.(((....)))-..........-...................((((((--....)))))).((((....))))..--........- ( -14.70, z-score = -0.04, R) >droPer1.super_1 2958450 79 - 10282868 ----------------------UGGACUUAUCC-UCUCUUCCAU-UAUGCCCAAUAUACCCCUUGGCUGC--CAGGGUAGCCGUAUCGUGCGAUAAA--AACUUUUC- ----------------------.(((....)))-..........-...................((((((--....)))))).((((....))))..--........- ( -14.40, z-score = -0.10, R) >droWil1.scaffold_180772 3425720 91 - 8906247 ----------------AAAUUCCUGCCUCAUCUAUAUGUGACUU-CUUAGGGGAUAUGCCCGUUCCUUGUGGUAGGGUAACCAUUUCGUGCGAUAAACUGAUUUUGCA ----------------....(((((((((((......))))...-..((((((((......)))))))).)))))))...........(((((..........))))) ( -20.10, z-score = -0.10, R) >consensus AAAAGCUAACCC_UCA__UUUGUUUGCUUAUCCAAUUAAAAU___AACAGCUUGAAAUUCCCUU_CAAAUUGAAGGGUAUCGCUGUCGUGCGAUAAA__UAUCUUGCC ..............................................(((((........((((..........))))....)))))...................... ( -4.37 = -4.26 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:24:00 2011