| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,936,863 – 11,936,955 |
| Length | 92 |
| Max. P | 0.989471 |

| Location | 11,936,863 – 11,936,955 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 73.78 |
| Shannon entropy | 0.52319 |
| G+C content | 0.37454 |
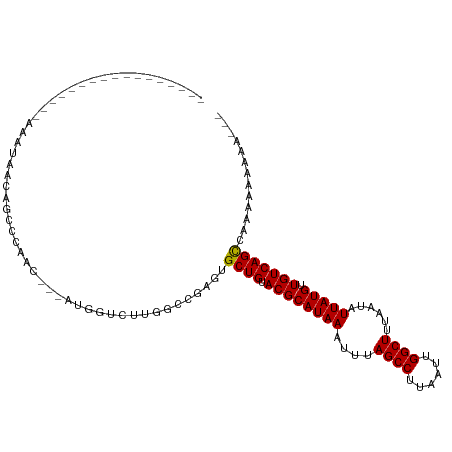
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -15.64 |
| Energy contribution | -15.39 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.989471 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
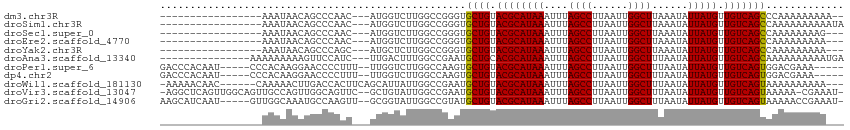
>dm3.chr3R 11936863 92 - 27905053 -----------------AAAUAACAGCCCAAC---AUGGUCUUGGCCGGGUGCUGUACGCAUAAAUUUAGCCUUAAUUGGCUUAAAUAUUAUGUUGUCAGCCCAAAAAAAAA-- -----------------...............---.((((....))))((.((((.((((((((((((((((......))).))))).))))).))))))))).........-- ( -25.20, z-score = -2.00, R) >droSim1.chr3R 17977525 94 + 27517382 -----------------AAAUAACAGCCCAAC---AUGGUCUUGGCCGGGUGCUGUACGCAUAAAUUUAGCCUUAAUUGGCUUAAAUAUUAUGUUGUCAGCCAAAAAAAAAAUA -----------------........((((...---..(((....)))))))((((.((((((((((((((((......))).))))).))))).)))))))............. ( -25.10, z-score = -2.09, R) >droSec1.super_0 11097693 91 + 21120651 -----------------AAAUAACAGCCCAAC---AUGGUCUUGGCCGGGUGCUGUACGCAUAAAUUUAGCCUUAAUUGGCUUAAAUAUUAUGUUGUCAGCCAAAAAAAAG--- -----------------........((((...---..(((....)))))))((((.((((((((((((((((......))).))))).))))).)))))))..........--- ( -25.10, z-score = -2.13, R) >droEre2.scaffold_4770 11460638 91 - 17746568 -----------------AAAUAACAGCCCAAC---AUGGUCUUGGCCGGGUGCUGUACGCAUAAAUUUAGCCUUAAUUGGCUUAAAUAUUAUGUUGUCAGCCAAAAAAAAA--- -----------------........((((...---..(((....)))))))((((.((((((((((((((((......))).))))).))))).)))))))..........--- ( -25.10, z-score = -2.07, R) >droYak2.chr3R 14334792 91 + 28832112 -----------------AAAUAACAGCCCAGC---AUGCUCUUGGCCGGGUGCUGUACGCAUAAAUUUAGCCUUAAUUGGCUUAAAUAUUAUGUUGUCAGCCAAAAAAAAA--- -----------------........((((.((---.........)).))))((((.((((((((((((((((......))).))))).))))).)))))))..........--- ( -25.20, z-score = -1.91, R) >droAna3.scaffold_13340 2060046 96 + 23697760 ---------------AAAAAAAAAGUUCCAUC---UUGACUUUGGCCGAAUGCUGCACGCAUAAAUUUAGCCUUAAUUGGCUUUAAUAUUAUGUUGUCAGCAAAAAAAAAAUGA ---------------......((((((.....---..)))))).......(((((((.(((((((((.((((......))))..))).)))))))).)))))............ ( -20.50, z-score = -1.79, R) >droPer1.super_6 4679370 102 + 6141320 GACCCACAAU-----CCCACAAGGAACCCCUUU--UUGGUCUUGGCCAAGUGCUGUACGCAUAAAUUUAGCCUUAAUUGGCUUUAAUAUUAUGUUGUCAGUGGACGAAA----- ...((((...-----..(((((((....)))).--(((((....)))))))).((.(((((((((((.((((......))))..))).))))).)))))))))......----- ( -26.70, z-score = -1.66, R) >dp4.chr2 4662967 102 + 30794189 GACCCACAAU-----CCCACAAGGAACCCCUUU--UUGGUCUUGGCCAAGUGCUGUACGCAUAAAUUUAGCCUUAAUUGGCUUUAAUAUUAUGUUGUCAGUGGACGAAA----- ...((((...-----..(((((((....)))).--(((((....)))))))).((.(((((((((((.((((......))))..))).))))).)))))))))......----- ( -26.70, z-score = -1.66, R) >droWil1.scaffold_181130 8754859 104 + 16660200 -AAAAACAAC------CAAAAACUUGACCACUUCAGCAUUAUUGGCCGAAUGCUGUACGCAUAAAUUUAGCCUUAAUUGGCUUUAAUAUUAUGUUGUCAGUAAAAAAAAAA--- -.........------.....(((.(((.....(((((((........)))))))...(((((((((.((((......))))..))).)))))).))))))..........--- ( -20.90, z-score = -2.14, R) >droVir3.scaffold_13047 15221222 109 + 19223366 -AGGCUCAGUUGGCAGUUGCCAGUUGGCAGUUC--GCUGUAUUGGCCGAAUGCUGUACGCAUAAAUUUAGCCUUAAUUGGCUUUAAUAUUAUGUUGUCAGUAAAAA-CGAAAU- -.((((((..((((....))))..))((((...--.))))...))))...(((((.(((((((((((.((((......))))..))).))))).))))))))....-......- ( -34.80, z-score = -2.51, R) >droGri2.scaffold_14906 13522683 106 + 14172833 AAGCAUCAAU-----GUUGGCAAAUGCCAAGUU--GCGGUAUUGGCCGUAUGCUGUACGCAUAAAUUUAGCCUUAAUUGGCUUUAAUAUUAUGUUGUCAGUAAAAACCGAAAU- ..........-----.(((((....)))))..(--(((((....))))))(((((.(((((((((((.((((......))))..))).))))).))))))))...........- ( -29.80, z-score = -1.99, R) >consensus _________________AAAUAACAGCCCAAC___AUGGUCUUGGCCGAGUGCUGUACGCAUAAAUUUAGCCUUAAUUGGCUUUAAUAUUAUGUUGUCAGCCAAAAAAAAA___ ...................................................((((.((((((((....((((......))))......))))).)))))))............. (-15.64 = -15.39 + -0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:18:57 2011