| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,936,693 – 11,936,790 |
| Length | 97 |
| Max. P | 0.511570 |

| Location | 11,936,693 – 11,936,790 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 69.22 |
| Shannon entropy | 0.59977 |
| G+C content | 0.52556 |
| Mean single sequence MFE | -29.36 |
| Consensus MFE | -12.87 |
| Energy contribution | -12.25 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.511570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
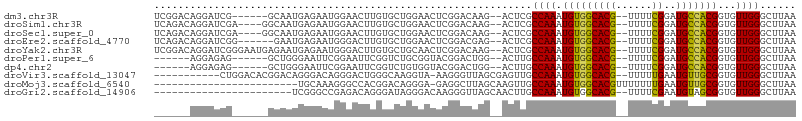
>dm3.chr3R 11936693 97 - 27905053 UCGGACAGGAUCG------GCAAUGAGAAUGGAACUUGUGCUGGAACUCGGACAAG--ACUCGCCAAAUGUGGCACG--UUUUCGGAUGCCACGGUGUUGGGCUUAA ..(..(((.((((------(((.((((((((...(((((.(((.....))))))))--....((((....)))).))--))))))..))))..))).)))..).... ( -30.60, z-score = -1.03, R) >droSim1.chr3R 17977347 99 + 27517382 UCAGACAGGAUCGA----GGCAAUGAGAAUGGAACUUGUGCUGGAACUCGGACAAG--ACUCGCCAAAUGUGGCACG--UUUUCGGAUGCCACGGUGUUGGGCUUAA .....(((.((((.----((((.((((((((...(((((.(((.....))))))))--....((((....)))).))--))))))..)))).)))).)))....... ( -34.80, z-score = -2.35, R) >droSec1.super_0 11097521 99 + 21120651 UCAGACAGGAUCGA----GGCAAUGAGAAUGGAACUUGUGCUGGAACUCGGACAAG--ACUCGCCAAAUGUGGCACG--UUUUCGGAUGCCACGGUGUUGGGCUUAA .....(((.((((.----((((.((((((((...(((((.(((.....))))))))--....((((....)))).))--))))))..)))).)))).)))....... ( -34.80, z-score = -2.35, R) >droEre2.scaffold_4770 11460473 97 - 17746568 UCAGACAGGAUCGG------GAAUGAGAAUGGGACUUGUGCUGGAACUCGGACGAG--ACUCGCCAAAUGUGGCACG--UUUUCGGAUGCCACGGUGUUGGGCUUAA .....(((.(((((------(..((((((((((.(((((.(((.....))))))))--.)))((((....))))..)--))))))....)).)))).)))....... ( -30.10, z-score = -0.77, R) >droYak2.chr3R 14334616 103 + 28832112 UCGGACAGGAUCGGGAAUGAGAAUGAGAAUGGGACUUGUGCUGCAACUCGGACAAG--ACUCGCCAAAUGUGGCACG--UUUUCGGAUGCCACGGUGUUGGGCUUAA ..(..(((.((((((..((((((((.....(((.(((((.(((.....))))))))--.)))((((....)))).))--))))))....)).)))).)))..).... ( -32.80, z-score = -1.24, R) >droPer1.super_6 4679218 91 + 6141320 ------AGGAGAG------GCUGGGAAUUCGGAAUUCGGUCUGCGGUACGGACUGG--ACUUGCCAAAUGUGGCACG--UUUUCGGAUGCCACGGUGUUGGGCUUAA ------(((.((.------((((((.(((((((((((((((((.....))))))))--)..(((((....)))))..--.)))))))).)).)))).))...))).. ( -32.80, z-score = -1.92, R) >dp4.chr2 4662815 91 + 30794189 ------AGGAGAG------GCUGGGAAUUCGGAAUUCGGUCUGUGGUACGGACUGG--ACUUGCCAAAUGUGGCACG--UUUUCGGAUGCCACGGUGUUGGGCUUAA ------(((.((.------((((((.(((((((((((((((((.....))))))))--)..(((((....)))))..--.)))))))).)).)))).))...))).. ( -33.60, z-score = -2.44, R) >droVir3.scaffold_13047 15221066 93 + 19223366 -----------CUGGACACGGACAGGGACAGGGACUGGGCAAGGUA-AAGGGUUAGCGAGUUGCCAAAUGUGGCACG--UUUUUGAAUGUUGCGGUGUUGGGCUUAA -----------((.(((((...(((.........))).((((.((.-.(((...((((...(((((....)))))))--)))))..)).)))).))))).))..... ( -22.30, z-score = 0.37, R) >droMoj3.scaffold_6540 13146043 82 + 34148556 ------------------------UGCAAAGGGCCACGGACAGGGA-GAGGCUUAGCAAGUUGCCAAAUGUGGCACGUUUUUUUGAAUGUUGCGGUGUUGGGCUUAA ------------------------(((...(((((.(.........-).))))).)))....(((.((..(.((((((((....))))).))).)..)).))).... ( -22.00, z-score = 0.34, R) >droGri2.scaffold_14906 13522532 82 + 14172833 -----------------------UCGGGCCGAGACAGGGAUAGGGACAAGGGUUAGCAACUUGCCAAAUGUGGCACG--UUUUCGAAUGUAGCGGUGUUGGGCUUAA -----------------------..((.((((.((....(((.((.((((.........))))))...))).(((((--((....))))).)).)).)))).))... ( -19.80, z-score = 0.57, R) >consensus ______AGGAUCG______GCAAUGAGAAUGGAACUUGGGCUGGAACUCGGACUAG__ACUCGCCAAAUGUGGCACG__UUUUCGGAUGCCACGGUGUUGGGCUUAA ...............................................................((((.(((((((((......))..)))))))...))))...... (-12.87 = -12.25 + -0.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:18:56 2011