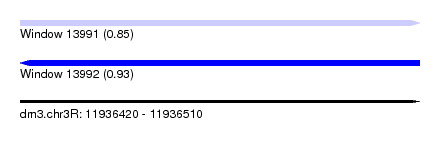
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,936,420 – 11,936,510 |
| Length | 90 |
| Max. P | 0.934239 |

| Location | 11,936,420 – 11,936,510 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 75.80 |
| Shannon entropy | 0.46597 |
| G+C content | 0.54999 |
| Mean single sequence MFE | -29.74 |
| Consensus MFE | -18.76 |
| Energy contribution | -18.10 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.851510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
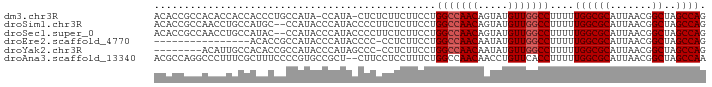
>dm3.chr3R 11936420 90 + 27905053 CUGGCUAGCCGUUAAUGCGCCAAAAAGGCCAACAUACUGUUGGCCAGGAAGAAGAGAG-UAUGG-UAUGGCAGGGUGGUGGUGUGGCGGUGU .......(((((((...(((((.....((((.((((((.((..(......)..)).))-)))).-..))))....)))))...))))))).. ( -31.90, z-score = -1.93, R) >droSim1.chr3R 17977078 90 - 27517382 CUGGCUAGCCGUUAAUGCGCCAAAAAGGCCAACAUACUGUUGGCCAGGAAGAGAAGGGGUAUGGGUAUGG--GCAUGGCAGGUUGGCGGUGU ...(((((((....((((.((.....(((((((.....))))))).............(((....)))))--))))....)))))))..... ( -31.90, z-score = -1.44, R) >droSec1.super_0 11097252 90 - 21120651 CUGGCUAGCCGUUAAUGCGCCAAAAAGGCCAACAUACUGUUGGCCAGGAAGAGAAGGGGUAUGGGUAUGG--GUAUGGCAGGUUGGCGGUGU ...(((((((((....))(((.....(((((((.....))))))).............((((........--))))))).)))))))..... ( -30.10, z-score = -1.23, R) >droEre2.scaffold_4770 11460221 75 + 17746568 CUGGCUAGCCGUUAAUGCGCCAAAAAGGCCAACAUAUUGUUGGCCAGGAAGAGG-GGGGUAUGGGUAUGGCGGUGU---------------- ...(((.(((((..((((.((.....(((((((.....))))))).........-)).))))....))))))))..---------------- ( -26.94, z-score = -1.80, R) >droYak2.chr3R 14334356 83 - 28832112 CUGGCUAGCCGUUAAUGCGCCAAAAAGGCCAACAUAUUGUUGGCCAGGAAGAGG-GGGCUAUGGGUAUGGCGGUGUGGCAAUGU-------- ...(((((((((((.(((.(((....(((((((.....)))))))......((.-...)).))))))))))))).)))).....-------- ( -30.70, z-score = -1.53, R) >droAna3.scaffold_13340 2059630 90 - 23697760 UUGGCUAGCCGUUAAUGCGCCAAAAAGGUGAACAGGUUGUUGGCCAGAAAGGAGGAAG--AGCGGCACGGGGAAAGCGAAAGGGCCUGGCGU ..((((.((((((..(.(.((.....(((.(((.....))).))).....)).).)..--))))))..........(....)))))...... ( -26.90, z-score = -0.38, R) >consensus CUGGCUAGCCGUUAAUGCGCCAAAAAGGCCAACAUACUGUUGGCCAGGAAGAGGAGGGGUAUGGGUAUGGCGGUAUGGCAGGGUGGCGGUGU (((((((((.((......(((.....)))......)).)))))))))............................................. (-18.76 = -18.10 + -0.66)

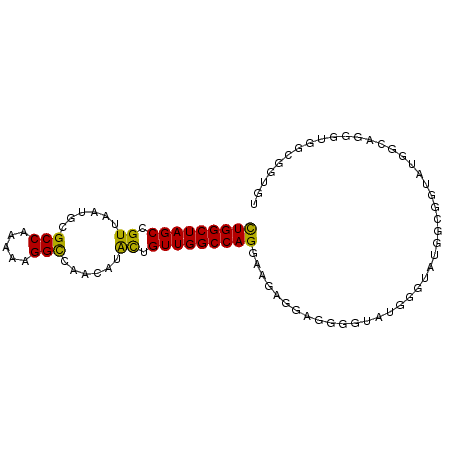

| Location | 11,936,420 – 11,936,510 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 75.80 |
| Shannon entropy | 0.46597 |
| G+C content | 0.54999 |
| Mean single sequence MFE | -19.72 |
| Consensus MFE | -16.20 |
| Energy contribution | -16.53 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11936420 90 - 27905053 ACACCGCCACACCACCACCCUGCCAUA-CCAUA-CUCUCUUCUUCCUGGCCAACAGUAUGUUGGCCUUUUUGGCGCAUUAACGGCUAGCCAG ....(((((...((......)).....-.....-.............(((((((.....)))))))....))))).......((....)).. ( -19.80, z-score = -1.39, R) >droSim1.chr3R 17977078 90 + 27517382 ACACCGCCAACCUGCCAUGC--CCAUACCCAUACCCCUUCUCUUCCUGGCCAACAGUAUGUUGGCCUUUUUGGCGCAUUAACGGCUAGCCAG ....((((((......(((.--.......)))...............(((((((.....)))))))...)))))).......((....)).. ( -21.90, z-score = -1.46, R) >droSec1.super_0 11097252 90 + 21120651 ACACCGCCAACCUGCCAUAC--CCAUACCCAUACCCCUUCUCUUCCUGGCCAACAGUAUGUUGGCCUUUUUGGCGCAUUAACGGCUAGCCAG ....((((((..((......--.))......................(((((((.....)))))))...)))))).......((....)).. ( -20.20, z-score = -1.45, R) >droEre2.scaffold_4770 11460221 75 - 17746568 ----------------ACACCGCCAUACCCAUACCCC-CCUCUUCCUGGCCAACAAUAUGUUGGCCUUUUUGGCGCAUUAACGGCUAGCCAG ----------------....(((((............-.........(((((((.....)))))))....))))).......((....)).. ( -19.61, z-score = -2.25, R) >droYak2.chr3R 14334356 83 + 28832112 --------ACAUUGCCACACCGCCAUACCCAUAGCCC-CCUCUUCCUGGCCAACAAUAUGUUGGCCUUUUUGGCGCAUUAACGGCUAGCCAG --------.....(((....(((((............-.........(((((((.....)))))))....))))).......)))....... ( -20.61, z-score = -1.27, R) >droAna3.scaffold_13340 2059630 90 + 23697760 ACGCCAGGCCCUUUCGCUUUCCCCGUGCCGCU--CUUCCUCCUUUCUGGCCAACAACCUGUUCACCUUUUUGGCGCAUUAACGGCUAGCCAA ..(((((((......)))).....(((((...--......((.....))..(((.....))).........)))))......)))....... ( -16.20, z-score = 0.82, R) >consensus ACACCGCCACCCUGCCACACCGCCAUACCCAUACCCCUCCUCUUCCUGGCCAACAAUAUGUUGGCCUUUUUGGCGCAUUAACGGCUAGCCAG ...............................................(((((((.....)))))))....((((((.......))..)))). (-16.20 = -16.53 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:18:56 2011