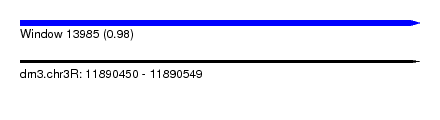
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,890,450 – 11,890,549 |
| Length | 99 |
| Max. P | 0.980269 |

| Location | 11,890,450 – 11,890,549 |
|---|---|
| Length | 99 |
| Sequences | 13 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 59.24 |
| Shannon entropy | 0.90067 |
| G+C content | 0.47001 |
| Mean single sequence MFE | -33.65 |
| Consensus MFE | -7.11 |
| Energy contribution | -7.57 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.21 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
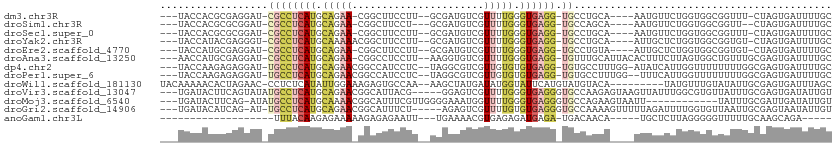
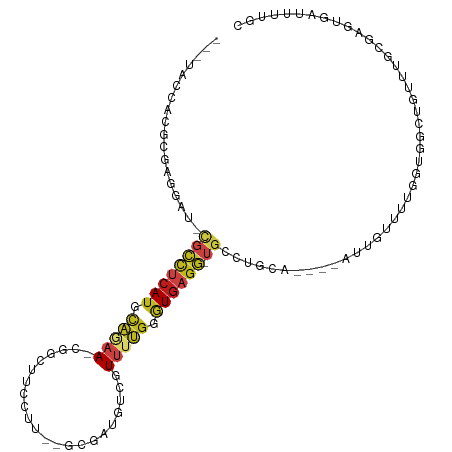
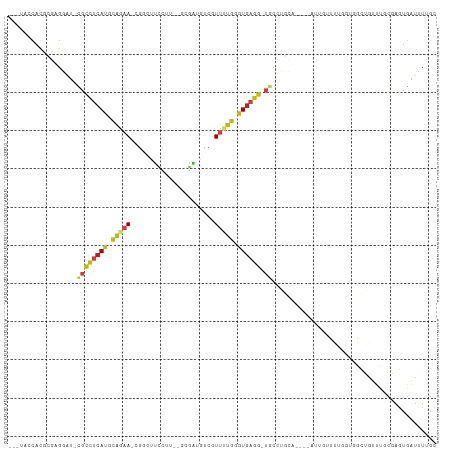
>dm3.chr3R 11890450 99 + 27905053 ---UACCACGCGAGGAU-CGCCUCAUGCAGAA-CGGCUUCCUU--GCGAUGUCGUUUUGGGUGAGG-UGCCUGCA----AAUGUUCUGGUGGCGGUUU-CUAGUGAUUUUGC ---.....(((.(((((-((((((((.(((((-((((.((...--..)).))))))))).))))))-))......----...))))).)))((((..(-(....))..)))) ( -37.50, z-score = -2.08, R) >droSim1.chr3R 17931556 97 - 27517382 ---UACCACGCGCGGAU-CGCCUCAUGCAGAA-CGGCUUCCU---GCGAUGUCGUUUUGGGUGAGG-UGCCAGCA----AAUGUUCUGGUGGCGGUU--CUAGUGAUUUUGC ---.....(((..((((-((((((((.(((((-((((.((..---..)).))))))))).))))))-)((((.((----.......)).))))))))--)..)))....... ( -40.90, z-score = -2.75, R) >droSec1.super_0 11051670 99 - 21120651 ---UACCACGCGCGGAU-CGCCUCAUGCAGAA-CGGCUUCCUU--GCGAUGUCGUUUUGGGUGAGG-UGCCUGCA----AAUGUUCUGGUGGCGGUUU-CUAGUGAUUUUGC ---(((((.((((((..-((((((((.(((((-((((.((...--..)).))))))))).))))))-)).)))).----...))..)))))((((..(-(....))..)))) ( -38.90, z-score = -2.24, R) >droYak2.chr3R 14287498 100 - 28832112 ---UACCAUACGAGGGU-CGCCUCAUGCAAAAACGGCUUCCUU--GCGAUGUCGUUUUGGGUGAGG-UGCCUGCA----AUUGCUCUGGUGGCGGUGU-CUAGUGAUUUUGC ---(((((.....(((.-((((((((.(((((.((((.((...--..)).))))))))).))))))-)))))((.----...))..)))))((((.((-(....))).)))) ( -37.00, z-score = -2.08, R) >droEre2.scaffold_4770 11412745 99 + 17746568 ---UACCAUGCGAGGAU-CGCCUCAUGCAGAA-CGGCUUCCUU--GCGAUGUCGUUUUGGGUGAGG-UGCCUGUA----AUUGCUCUGGUGGCGGUGU-CUAGUGAUUUUGC ---(((((.((((..((-((((((((.(((((-((((.((...--..)).))))))))).))))))-))...)).----.))))..)))))((((.((-(....))).)))) ( -39.80, z-score = -2.91, R) >droAna3.scaffold_13250 299923 104 + 3535662 ---AACCAUGCGAGGAU-CGCCUCAUGCAGAA-CGGCCUCCUU--AAGGUGUCGUUUUGGGUGAGG-UGUUUGCAUUACACUUUCUUAGUGGCUGUUUGCGAGUGAUUUUGC ---......((((((..-((((((((.(((((-((((..((..--..)).))))))))).))))))-))((((((...((((.....))))......))))))...)))))) ( -39.70, z-score = -3.31, R) >dp4.chr2 4608889 104 - 30794189 ---UACCAAGAGAGGAU-UGCCUCAUGCAGAACGGCCAUCCUC--UAGGCGUCGUUGUGUGUGAGG-UGUGCCUUUGG-AUAUCAUUGGUUUUUUUUGGCGAGUGAUUUUGC ---..(((...((((..-.((((((((((.((((((...((..--..)).)))))).)))))))))-)...)))))))-..((((((.(((......))).))))))..... ( -39.40, z-score = -3.52, R) >droPer1.super_6 4624937 103 - 6141320 ---UACCAAGAGAGGAU-UGCCUCAUGCAGAACGGCCAUCCUC--UAGGCGUCGUUGUGUGUGAGG-UGUGCCUUUGG--UUUCAUUGGUUUUUUUUGGCGAGUGAUUUUGC ---.((((...((((..-.((((((((((.((((((...((..--..)).)))))).)))))))))-)...)))))))--).(((((.(((......))).)))))...... ( -39.20, z-score = -3.57, R) >droWil1.scaffold_181130 8700017 100 - 16660200 UACAAAAACACUAGAAC-CCUCUCAUAUUGGAAAGAGUGCCAA--AAGCUAUGAUAUGGUAUUCAUGUAUGUACA---------UAUGUUUGUAUAUUGCGAGUGAUUUAGC ..........(((((.(-.(((............((((((((.--...........))))))))..(((((((((---------......)))))).)))))).).))))). ( -21.00, z-score = -1.04, R) >droVir3.scaffold_13047 15164004 104 - 19223366 ---UGAUACUUCAGUAUAUGCCUCAUGCAGAACGGCAUUACG-----GGAGUCGUUUUGGGUGAGGGUGCCAAGAGUAAGUUAUUUGGCGUGUUAUUUGCGAGUGAUAUUGU ---...((((((((.((((.((((((.(((((((((......-----...))))))))).))))))..((((((.(.....).))))))))))...))).)))))....... ( -32.80, z-score = -2.56, R) >droMoj3.scaffold_6540 13093979 96 - 34148556 ---UGAUACUUCAG-AUAUGCCUCAUGCAAAACGGCAUUUCGUUGGGGAAAUGGUUUUGGGUGAGGGUGCCAGAAGUAAUU------------UAUUUGCGAUUGAUAUUGU ---...((((((.(-.(((.((((((.((((((..((((((......)))))))))))).))))))))).).))))))...------------.....(((((....))))) ( -28.70, z-score = -2.49, R) >droGri2.scaffold_14906 13466559 102 - 14172833 ---UGAUACAUCAG-AU-UGCCUCAUGCAGAACGGCAUUUCU-----AGAGUCGUUUUGUGUGAGGGUGCCAAAAGUUUUUAGAUUUUGGUGUUAAUUGCGAGUAAUAUUGU ---...(((.((.(-((-((((((((((((((((((......-----...))))))))))))))))(..((((((.........))))))..))))))..)))))....... ( -33.20, z-score = -3.88, R) >anoGam1.chr3L 17274039 79 + 41284009 -------------------UUUACAAGAGAAAAAGAGAGAAUU---UGAAAACGUGAGAGAUGAGA-UGACAACA-----UGCUCUUAGGGGGUUUUUGCAAGCAGA----- -------------------...............(..((((((---(.....(.(((((((((...-......))-----).)))))).))))))))..).......----- ( -9.30, z-score = 0.72, R) >consensus ___UACCACGCGAGGAU_CGCCUCAUGCAGAA_CGGCUUCCUU__GCGAUGUCGUUUUGGGUGAGG_UGCCUGCA____AUUGUUUUGGUGGCUGUUUGCGAGUGAUUUUGC ....................((((((.(((((......................))))).)))))).............................................. ( -7.11 = -7.57 + 0.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:18:50 2011