| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,862,274 – 11,862,367 |
| Length | 93 |
| Max. P | 0.935464 |

| Location | 11,862,274 – 11,862,367 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 70.44 |
| Shannon entropy | 0.57224 |
| G+C content | 0.53514 |
| Mean single sequence MFE | -29.32 |
| Consensus MFE | -15.10 |
| Energy contribution | -14.67 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.935464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
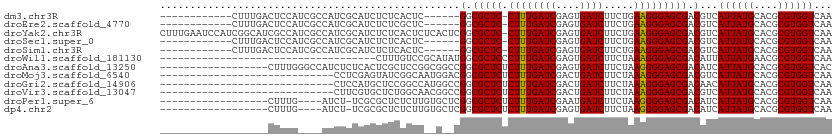
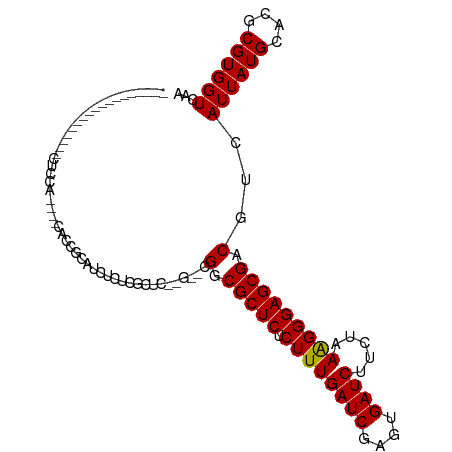
>dm3.chr3R 11862274 93 + 27905053 ------------CUUUGACUCCAUCGCCAUCGCAUCUCUCACUC------GGCGCUC-CUUUGAUCGAGUGAUCUUCUGAAGGGAGCGACGUCAUUAUGCACGCGUGGUCAA ------------..((((((....(((....((((........(------(.(((((-((((((((....)))).....))))))))).)).....))))..))).)))))) ( -27.32, z-score = -0.58, R) >droEre2.scaffold_4770 11384435 93 + 17746568 ------------CUUUGACUCCAUCGCCAUCGCAUCUCUCGCUC------GGCGCUC-CUUUGAUCGAGUGAUCUUCUGAAGGGAGCGACGUCAUUAUGCACGCGUGGUCAA ------------..((((((....(((....((((........(------(.(((((-((((((((....)))).....))))))))).)).....))))..))).)))))) ( -27.32, z-score = -0.12, R) >droYak2.chr3R 14259342 111 - 28832112 CUUUGAAUCCAUCGGCAUCGCCAUCGCCAUCGCAUCUCUCACUCUCACUCGGCGCUC-CUUUGAUCGAGUGAUCUUCUGAAGGGAGCGACGUCAUUAUGCACGCGUGGUCAA ..((((..((((((((((......((...((((.(((((((...((((((((((...-...)).)))))))).....)).))))))))))).....)))).)).)))))))) ( -31.00, z-score = -0.15, R) >droSec1.super_0 11023818 93 - 21120651 ------------CUUUGACUCCAUCGCCAUCGCAUCUCUCACUC------GGCGCUC-CUUUGAUCGAGUGAUCUUCUGAAGGGAGCGACGUCAUUAUGCACGCGUGGUCAA ------------..((((((....(((....((((........(------(.(((((-((((((((....)))).....))))))))).)).....))))..))).)))))) ( -27.32, z-score = -0.58, R) >droSim1.chr3R 17903027 93 - 27517382 ------------CUUUGACUCCAUCGCCAUCGCAUCUCUCACUC------GGCGCUC-CUUUGAUCGAGUGAUCUUCUGAAGGGAGCGACGUCAUUAUGCACGCGUGGUCAA ------------..((((((....(((....((((........(------(.(((((-((((((((....)))).....))))))))).)).....))))..))).)))))) ( -27.32, z-score = -0.58, R) >droWil1.scaffold_181130 8649941 76 - 16660200 ------------------------------------CUUUGUCCGCAUAUGGCGCUCCCUUUGAUCGAGUGAUCUUCUAAAGGGAGCGACAUUAUUAUGAACGCGUGGUCAA ------------------------------------..(((.((((......(((((((((((...((....))...))))))))))).(((....))).....)))).))) ( -27.80, z-score = -3.39, R) >droAna3.scaffold_13250 270532 94 + 3535662 ------------------CUUUGGGCCAUCUCUCACUCGCUCCGGCGGCCGGCGCUCUCUUUGAUCGAGUGAUCUUCUAAGGGGAGCGACAUCAUUAUGCACGCGUGGUCAC ------------------.....(((((.........(((.((((...))))(((((((((((...((....))...)))))))))))..............)))))))).. ( -28.80, z-score = -0.06, R) >droMoj3.scaffold_6540 13063354 83 - 34148556 -----------------------------CCUCGAGUAUCGGCAAUGGACGGCGCUCUCUUUGAUCGACUGAUCUUCUAAAGGGAGCGACGUCAUUAUGCACGCGUGGUCAA -----------------------------....((.((((((((...((((.((((((((((((((....))))....)))))))))).))))....))).)).))).)).. ( -31.70, z-score = -2.78, R) >droGri2.scaffold_14906 13435816 83 - 14172833 -----------------------------CUCCAUGCUCCGGCCAUGGCCGGCGCUCUCUUUGAUCGACUGAUCUUCUAAAGGGAGCGACAACAUUAUGCACGCGUGGUCAA -----------------------------..((((((.(((((....)))))((((((((((((((....))))....))))))))))..............)))))).... ( -35.80, z-score = -4.36, R) >droVir3.scaffold_13047 15131289 83 - 19223366 -----------------------------CUUCGUGCUCUGGCAACGGCCGGCGCUCUCUUUGAUCGACUGAUCUUCUAAAGGGAGCGACGUCAUUAUGCACGCGUGGUCAA -----------------------------...(((((.(((((....)))))((((((((((((((....))))....))))))))))..........)))))......... ( -30.10, z-score = -2.12, R) >droPer1.super_6 4596872 89 - 6141320 ------------------CUUUG----AUCU-UCGCGCUCUCUUGUGCUCGGCGCUCUCUUUGAUCGAAUGAUCUUCUAAGGGGAGCGACAUCAUUAUGCACGCGUGGUCAA ------------------..(((----(((.-.((((..(....(((.....((((((((((((((....))))....))))))))))....)))...)..)))).)))))) ( -28.50, z-score = -1.89, R) >dp4.chr2 4579412 89 - 30794189 ------------------CUUUG----AUCU-UCGCGCUCUCUUGUGCUCGGCGCUCUCUUUGAUCGAGUGAUCUUCUAAGGGGAGCGACAUCAUUAUGCACGCGUGGUCAA ------------------..(((----(((.-.((((..(....(((.....(((((((((((...((....))...)))))))))))....)))...)..)))).)))))) ( -28.90, z-score = -1.48, R) >consensus __________________CUCCA____CACCGCAUCUCUCGCUC__G__CGGCGCUCUCUUUGAUCGAGUGAUCUUCUAAAGGGAGCGACGUCAUUAUGCACGCGUGGUCAA ..................................................(.(((((.((((((((....))))....)))).))))).)...((((((....))))))... (-15.10 = -14.67 + -0.43)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:18:47 2011