
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,835,442 – 11,835,596 |
| Length | 154 |
| Max. P | 0.659907 |

| Location | 11,835,442 – 11,835,536 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 70.94 |
| Shannon entropy | 0.52470 |
| G+C content | 0.44962 |
| Mean single sequence MFE | -27.49 |
| Consensus MFE | -14.98 |
| Energy contribution | -17.56 |
| Covariance contribution | 2.58 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.578424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
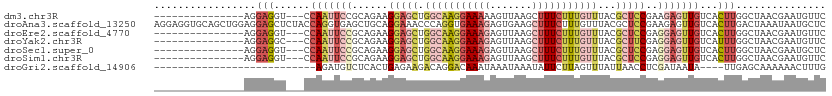
>dm3.chr3R 11835442 94 - 27905053 ---------------AGGAGGU---CCAAUUCCGCAGAAGGAGCUGGCAAGGAAAAAGUUAAGCUUUCUUUGUUUACGCUCCGAAGAGUUGUCACUUGGCUAACGAAUGUUC ---------------.((((..---....))))(((...(((((.((((((((((.........))))))))))...))))).....((((((....))).)))...))).. ( -25.10, z-score = -0.29, R) >droAna3.scaffold_13250 241437 112 - 3535662 AGGAGGUGCAGCUGGAGGAGCUCUACCAGGUGAGCUGCAGGAAACCCAGGUGAAAGAGUGAAGCUUUCUUUGUUUACGCUCCGAAGAGUUGUCACUUGACUAAAUAAUGCUC .(((((((((.((((.(.(((((........))))).).(....))))).))((((((.......))))))...))).))))...((((.(((....)))........)))) ( -32.80, z-score = 0.06, R) >droEre2.scaffold_4770 11357183 94 - 17746568 ---------------AGGAGGU---CCAAUUCCGCAGAAGGAGCUGGCAAGGAAAGAGUUAAGCUUUCUUUGUUUACGCUCCGAGGAGUUGUCACUUGGCUAACGAAUGUUC ---------------.(..(((---((((((((......(((((.(((((((((((.......)))))))))))...)))))..)))))))......))))..)........ ( -31.00, z-score = -1.67, R) >droYak2.chr3R 14231387 94 + 28832112 ---------------AGGAGGC---CCAAUUCCGCAGAAGGAGCUGGCAAGGAAAGAGUUAAGCUUUCUUUGUUUACGCUUCGAGGAGUUGUCAUUUGGCUAACGAAUGUUC ---------------.(..(((---((((((((...((((..(..(((((((((((.......)))))))))))..).))))..)))))))......))))..)........ ( -30.80, z-score = -1.82, R) >droSec1.super_0 10997073 94 + 21120651 ---------------AGGAGGU---CCAAUUCCGCAGAAGGAGCUGGCAAGGAAAGAGUUAAGCUUUCUUUGUUUACGCUCCGAGGAGUUGUCACUUGGCUAACGAAUGCUC ---------------..(((..---.(((((((......(((((.(((((((((((.......)))))))))))...)))))..)))))))...)))(((........))). ( -32.30, z-score = -1.72, R) >droSim1.chr3R 17876200 94 + 27517382 ---------------AGGAGGU---CCAAUUCCGCAGAAGGAGCUGGCAAGGAAAGAGUUAAGCUUUCUUUGUUUACGCUCCGAGGAGUUGUCACUUGGCUAACGAAUGUUC ---------------.(..(((---((((((((......(((((.(((((((((((.......)))))))))))...)))))..)))))))......))))..)........ ( -31.00, z-score = -1.67, R) >droGri2.scaffold_14906 13402679 81 + 14172833 ---------------------------AGAUGUCUCACUGAGAAGACAGGACAAAUAAAUAAAUAUUCUUAGUUUAUUAACCUCGAUAAUA----UUGAGCAAAAAACUUUG ---------------------------...((.(((((((((((.....................))))))))(((((......)))))..----..))))).......... ( -9.40, z-score = 0.00, R) >consensus _______________AGGAGGU___CCAAUUCCGCAGAAGGAGCUGGCAAGGAAAGAGUUAAGCUUUCUUUGUUUACGCUCCGAGGAGUUGUCACUUGGCUAACGAAUGUUC ..........................(((((((......(((((.(((((((((((.......)))))))))))...)))))..)))))))..................... (-14.98 = -17.56 + 2.58)


| Location | 11,835,503 – 11,835,596 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 67.19 |
| Shannon entropy | 0.57289 |
| G+C content | 0.56133 |
| Mean single sequence MFE | -32.81 |
| Consensus MFE | -16.14 |
| Energy contribution | -16.28 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.659907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
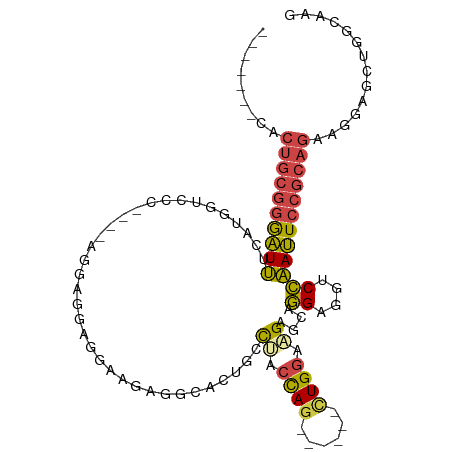
>dm3.chr3R 11835503 93 - 27905053 -------CACUGCGGGAUUUCAUGGUCCC----AGGUGGAGGAAGAGGCACUGCCUACCAG------CUGGAAGAGCAGGAGGUCCAAUUCCGCAGAAGGAGCUGGCAAG -------...(((.(((((((.((.((((----((.((((((.((.....)).))).))).------))))..)).)).)))))))..((((......))))...))).. ( -34.70, z-score = -1.14, R) >droEre2.scaffold_4770 11357244 97 - 17746568 -------CACUGCGGAAUUCCCGGGAUCCCAGGAGGAGGAGGAGGAGGUACUGCUUGCCAG------CUGGAAGAUCAGGAGGUCCAAUUCCGCAGAAGGAGCUGGCAAG -------..(((((((((((((..((((((((..((..(((.((......)).))).))..------))))..))))..).))...)))))))))).............. ( -34.10, z-score = -0.33, R) >droYak2.chr3R 14231448 97 + 28832112 -------CACUGCGGGAUUUCCCGGGUCC------CAGGAGGAGGAGAUACUGCCUACCAGAUCUGCUUGGAAGAUCAGGAGGCCCAAUUCCGCAGAAGGAGCUGGCAAG -------..((((((((((....((((((------(.(((((((......)).))).)).(((((.......))))).)).))))))))))))))).............. ( -38.60, z-score = -1.55, R) >droSec1.super_0 10997134 93 + 21120651 -------CACUGCGGGAUUUCAUGGUCCC----AGCAGGAGGUAGAGGCACUGCCAACCAG------CUGGAAGAGCAGGAGGUCCAAUUCCGCAGAAGGAGCUGGCAAG -------...(((.(((((((.((.((((----(((.((.(((((.....)))))..)).)------))))..)).)).)))))))..((((......))))...))).. ( -35.30, z-score = -1.65, R) >droSim1.chr3R 17876261 100 + 27517382 CACUCCGCACUGCGGGAUUUCAUGGUCCC----AGGAGGAGGUAGAGGCACUGCCCACCAG------CUGGAAGAGCAGGAGGUCCAAUUCCGCAGAAGGAGCUGGCAAG ((((((...((((((((((...((.((((----((..((.(((((.....)))))..))..------))))..)).))((....))))))))))))..)))).))..... ( -38.90, z-score = -1.12, R) >droGri2.scaffold_14906 13402732 77 + 14172833 ---------------GGUGACGUGAGUC-------AGUUAUAAAAAUGAAUUUACGAGUAA------CUGACCGAA-AAGAUGUCUCACUGAGAAGACAGGACAAA---- ---------------((((((....)))-------((((((................))))------)).)))...-....(((((..((....))...)))))..---- ( -15.29, z-score = -0.92, R) >consensus _______CACUGCGGGAUUUCAUGGUCCC____AGGAGGAGGAAGAGGCACUGCCUACCAG______CUGGAAGAGCAGGAGGUCCAAUUCCGCAGAAGGAGCUGGCAAG .........((((((((((...................................((.((((......)))).))....((....)))))))))))).............. (-16.14 = -16.28 + 0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:18:43 2011