| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,597,735 – 7,597,836 |
| Length | 101 |
| Max. P | 0.954229 |

| Location | 7,597,735 – 7,597,836 |
|---|---|
| Length | 101 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 76.57 |
| Shannon entropy | 0.45608 |
| G+C content | 0.48925 |
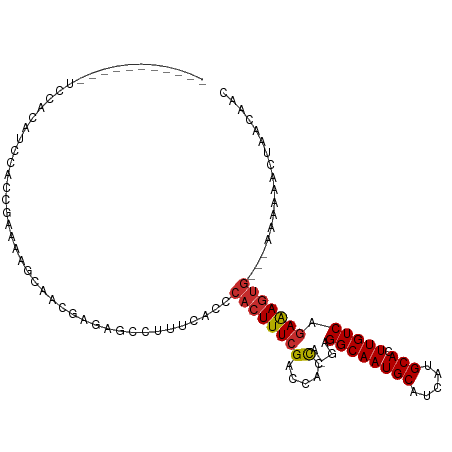
| Mean single sequence MFE | -22.37 |
| Consensus MFE | -12.19 |
| Energy contribution | -12.37 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.954229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
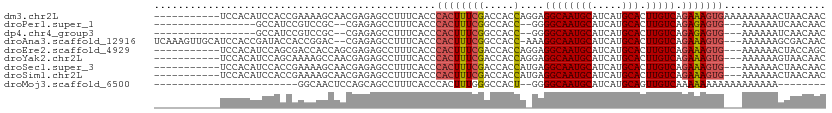
>dm3.chr2L 7597735 101 - 23011544 -----------UCCACAUCCACCGAAAAGCAACGAGAGCCUUUCACCCACUUUCGACCACCAGGAGGCAAUGCAUCAUGCACUUGUCAGAAAGUGAAAAAAAAACUAACAAC -----------............((((.((.......)).))))...(((((((..((....)).((((((((.....))).))))).)))))))................. ( -22.10, z-score = -2.94, R) >droPer1.super_1 2953006 88 - 10282868 -----------------GCCAUCCGUCCGC--CGAGAGCCUUUCACCCACUUUCGGCCACC--GGGGCAAUGCAUCAUGCACUUGUCAGAGAGUG---AAAAAAUCAACAAC -----------------(((..(((...((--((((((...........))))))))...)--)))))...........(((((......)))))---.............. ( -23.70, z-score = -1.39, R) >dp4.chr4_group3 1484587 88 - 11692001 -----------------GCCAUCCGUCCGC--CGAGAGCCUUUCACCCACUUUCGGCCACC--GGGGCAAUGCAUCAUGCACUUGUCAGAGAGUG---AAAAAAUCAACAAC -----------------(((..(((...((--((((((...........))))))))...)--)))))...........(((((......)))))---.............. ( -23.70, z-score = -1.39, R) >droAna3.scaffold_12916 15528474 106 - 16180835 UCAAAGUUGCAUCCACCGAUACCACCGGAC--CGAGAGCCUUUCACCCACUUUCGGCCACC-AAAGGCAAUGCAUCAUGCACUUGUCAGAAAGUG---AAAAAAGCGACAAC .....((((((((....)))..(((.((.(--((((((...........)))))))))...-...((((((((.....))).))))).....)))---......)))))... ( -28.30, z-score = -2.94, R) >droEre2.scaffold_4929 16516956 98 - 26641161 -----------UCCACAUCCAGCGACCACCAGCGAGAGCCUUUCACCCACUUUCGACCACCAGGAGGCAAUGCAUCAUGCACUUGUCAGAAAGUG---AAAAAACUACCAGC -----------..........((........((....))........(((((((..((....)).((((((((.....))).))))).)))))))---............)) ( -21.90, z-score = -1.92, R) >droYak2.chr2L 17026353 98 + 22324452 -----------UCCACAUCCAGCAAAAGCCAACGAGAGCCUUUCACCCACUUUCGACCACCAGGAGGCAAUGCAUCAUGCACUUGUCAGAAAGUG---AAAAAAGUAACAAC -----------..(((.(((.((....))...((((((...........)))))).......)))((((((((.....))).))))).....)))---.............. ( -19.70, z-score = -1.62, R) >droSec1.super_3 3111303 98 - 7220098 -----------UCCACAUCCACCGAAAAGCAACGAGAGCCUUUCACCCACUUUCGACCACCAUGAGGCAAUGCAUCAUGCACUUGUCAGAAAGUG---AAAAAACUAACAAC -----------............((((.((.......)).))))...((((((((((...(((((.((...)).))))).....))).)))))))---.............. ( -20.30, z-score = -2.69, R) >droSim1.chr2L 7393443 98 - 22036055 -----------UCCACAUCCACCGAAAAGCAACGAGAGCCUUUCACCCACUUUCGACCACCAUGAGGCAAUGCAUCAUGCACUUGUCAGAAAGUG---AAAAAACUAACAAC -----------............((((.((.......)).))))...((((((((((...(((((.((...)).))))).....))).)))))))---.............. ( -20.30, z-score = -2.69, R) >droMoj3.scaffold_6500 19808567 78 - 32352404 ------------------------GGCAACUCCAGCAGCCUUUCACCCACUUUGGGCCACU--GGGGCAAUGCAUCAUGCAGUUGUCAAAAAAAAAAAAAAAAA-------- ------------------------(((((((((((..((((............))))..))--)))....(((.....))))))))).................-------- ( -21.30, z-score = -1.24, R) >consensus ___________UCCACAUCCACCGAAAAGCAACGAGAGCCUUUCACCCACUUUCGACCACCA_GAGGCAAUGCAUCAUGCACUUGUCAGAAAGUG___AAAAAACUAACAAC ...............................................((((((((.....)....((((((((.....))).))))).)))))))................. (-12.19 = -12.37 + 0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:23:57 2011