
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,785,235 – 11,785,349 |
| Length | 114 |
| Max. P | 0.724958 |

| Location | 11,785,235 – 11,785,349 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.01 |
| Shannon entropy | 0.21662 |
| G+C content | 0.29661 |
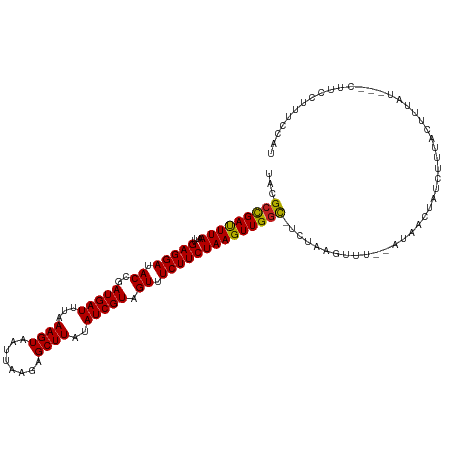
| Mean single sequence MFE | -20.89 |
| Consensus MFE | -19.18 |
| Energy contribution | -19.03 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.724958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
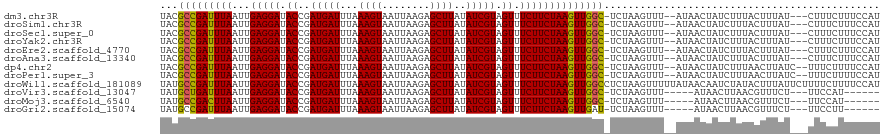
>dm3.chr3R 11785235 114 + 27905053 UACGCCGAUUUAAUUGAGGAUACCGAUGAUUUAAAGUAAUUAAGAGCUUAUAUCGUAGUUUCUUCUAAGUUGGC-UCUAAGUUU--AUAACUAUCUUUACUUUAU---CUUUCUUUCCAU ...(((((((((...(((((.((..(((((...((((........))))..))))).)).))))))))))))))-.........--...................---............ ( -20.90, z-score = -1.05, R) >droSim1.chr3R 17823878 114 - 27517382 UACGCCGAUUUAAUUGAGGAUACCGAUGAUUUAAAGUAAUUAAGAGCUUAUAUCGUAGUUUCUUCUAAGUUGGC-UCUAAGUUU--AUAACUAUCUUUACUUUAU---CUUUCUUUCCAU ...(((((((((...(((((.((..(((((...((((........))))..))))).)).))))))))))))))-.........--...................---............ ( -20.90, z-score = -1.05, R) >droSec1.super_0 10947043 114 - 21120651 UACGCCGAUUUAAUUGAGGAUACCGAUGAUUUAAAGUAAUUAAGAGCUUAUAUCGUAGUUUCUUCUAAGUUGGC-UCUAAGUUU--AUAACUAUCUUUACUUUAU---CUUUCUUUCCAU ...(((((((((...(((((.((..(((((...((((........))))..))))).)).))))))))))))))-.........--...................---............ ( -20.90, z-score = -1.05, R) >droYak2.chr3R 14180374 114 - 28832112 UACGCCGAUUUAAUUGAGGAUACCGAUGAUUUAAAGUAAUUAAGAGCUUAUAUCGUAGUUUCUUCUAAGUUGGC-UCUAAGUUU--AUAACUAUCUUUACUUUAU---CUUUCUUUCCAU ...(((((((((...(((((.((..(((((...((((........))))..))))).)).))))))))))))))-.........--...................---............ ( -20.90, z-score = -1.05, R) >droEre2.scaffold_4770 11306836 114 + 17746568 UACGCCGAUUUAAUUGAGGAUACCGAUGAUUUAAAGUAAUUAAGAGCUUAUAUCGUAGUUUCUUCUAAGUUGGC-UCUAAGUUU--AUAACUAUCUUUACUUUAU---CUUUCUUUCCAU ...(((((((((...(((((.((..(((((...((((........))))..))))).)).))))))))))))))-.........--...................---............ ( -20.90, z-score = -1.05, R) >droAna3.scaffold_13340 16390135 114 + 23697760 UACGCCGAUUUAAUUGAGGAUACCGAUGAUUUAAAGUAAUUAAGAGCUUAUAUCGUAGUUUCUUCUAAGUUGGC-UCUAAGUUU--AUAACUAUCUUUACUUUAU---CUUUCUUUCCAU ...(((((((((...(((((.((..(((((...((((........))))..))))).)).))))))))))))))-.........--...................---............ ( -20.90, z-score = -1.05, R) >dp4.chr2 20483795 115 + 30794189 UACGCCGAUUUAAUUGAGGAUACCGAUGAUUUAAAGUAAUUAAGAGCUUAUAUCGUAGUUUCUUCUAAGUUGGC-UCUAAGUUU--AUAACUAUCUUUAACUUAUC--UUUCUUUUCCAU ...(((((((((...(((((.((..(((((...((((........))))..))))).)).))))))))))))))-..((((((.--............))))))..--............ ( -22.52, z-score = -1.57, R) >droPer1.super_3 3232359 115 + 7375914 UACGCCGAUUUAAUUGAGGAUACCGAUGAUUUAAAGUAAUUAAGAGCUUAUAUCGUAGUUUCUUCUAAGUUGGC-UCUAAGUUU--AUAACUAUCUUUAACUUAUC--UUUCUUUUCCAU ...(((((((((...(((((.((..(((((...((((........))))..))))).)).))))))))))))))-..((((((.--............))))))..--............ ( -22.52, z-score = -1.57, R) >droWil1.scaffold_181089 7270946 120 - 12369635 UAUGCCGAUUUAAUUGAGGAUACCGAUGAUUUAAAGUAAUUAAGAGCUUAUAUCGUAGUUUCUUCUAAGUUGGCCUCUAAGUUUUUAUAACAAUCUAUACUUUAUUCUUUUCUUUUCCAU ...(((((((((...(((((.((..(((((...((((........))))..))))).)).))))))))))))))......(((.....)))............................. ( -20.40, z-score = -0.95, R) >droVir3.scaffold_13047 2459037 105 + 19223366 UAUGCUGAUUUAAUUGAGGAUACCGAUGAUUUAAAGUAAUUAAGAGCUUAUAUCGUAGUUUCUUCUAAGUUGGC-UCUAAGUUU-----AUAACUUAACGUUUCU---UUCCAU------ ...((..(((((...(((((.((..(((((...((((........))))..))))).)).))))))))))..))-..(((((..-----...)))))........---......------ ( -19.70, z-score = -0.89, R) >droMoj3.scaffold_6540 18956948 105 - 34148556 UAUGCCGACUUAAUUGAGGAUACCGAUGAUUUAAAGUAAUUAAGAGCUUAUAUCGUAGUUUCUUCUAAGUUGGC-UCUAAGUUU-----AUAACUUAACGUUUCU---UUCCAU------ ...(((((((((...(((((.((..(((((...((((........))))..))))).)).))))))))))))))-..(((((..-----...)))))........---......------ ( -24.50, z-score = -2.50, R) >droGri2.scaffold_15074 6033713 105 + 7742996 UAUGCCGAUUUAAUUGAGGAUACCGAUGAUUUAAAGUAAUUAAGAGCUUAUAUCGUAGUUUCUUCUAAGUUGAU-UCUAAGUUU-----AUAACUUAACGUUUCU---UUCCUU------ .....(((((((...(((((.((..(((((...((((........))))..))))).)).))))))))))))..-..(((((..-----...)))))........---......------ ( -15.70, z-score = 0.13, R) >consensus UACGCCGAUUUAAUUGAGGAUACCGAUGAUUUAAAGUAAUUAAGAGCUUAUAUCGUAGUUUCUUCUAAGUUGGC_UCUAAGUUU__AUAACUAUCUUUACUUUAU___CUUCCUUUCCAU ...(((((((((...(((((.((..(((((...((((........))))..))))).)).)))))))))))))).............................................. (-19.18 = -19.03 + -0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:18:38 2011