| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,771,375 – 11,771,474 |
| Length | 99 |
| Max. P | 0.965846 |

| Location | 11,771,375 – 11,771,474 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 64.34 |
| Shannon entropy | 0.67455 |
| G+C content | 0.56058 |
| Mean single sequence MFE | -33.34 |
| Consensus MFE | -11.97 |
| Energy contribution | -15.84 |
| Covariance contribution | 3.87 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.965846 |
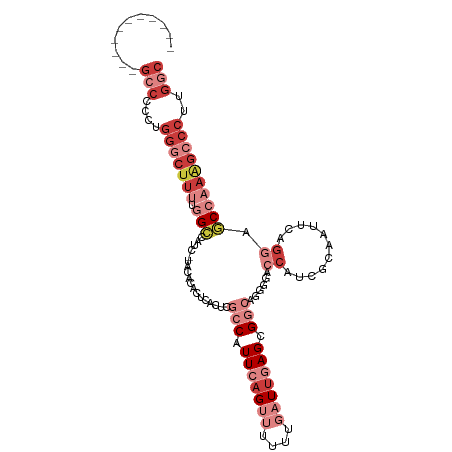
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11771375 99 + 27905053 -----------GCCCCCUGGGCUUUUGGCCAUCAUACACACUCACUCGCCAUUCAGUUUUUUGAUUGAGCGGCAGAGAACCAUCGCGAUUCAGGAGCCAAAGCCCUUGGC -----------(((....(((((((.(((...............((((((.(((((((....))))))).))).)))..((...........)).))))))))))..))) ( -33.40, z-score = -2.40, R) >droSim1.chr3R 17810173 99 - 27517382 -----------GCCUCCUGGGCUUUUGGCCAUCAUACACACUCACGCGCCAUUCAGUUUUUUGAUUGAGCGGCAGCGGACCAUCGCAAUUCAGGAGCCAAAGCCCUUGGC -----------(((....(((((((.(((............((.((((((.(((((((....))))))).))).)))))((...........)).))))))))))..))) ( -35.80, z-score = -2.54, R) >droSec1.super_0 10933268 99 - 21120651 -----------GCCCCCUGGGCUUUCGGCCAUCAUACACACUCACGCGCCAUUCAGUUUUUUGAUUGAGCGGCAGCGGACCAUCGCAAUUCAGGAGCCAAAGCCCUUGGC -----------(((....(((((((.(((............((.((((((.(((((((....))))))).))).)))))((...........)).))))))))))..))) ( -35.10, z-score = -2.28, R) >droYak2.chr3R 14166401 89 - 28832112 -----------GCCCCCUGGGCUUUUGGC--------ACACGCACUCGCCAUUCAGUUUUUUGAUUGAGCGGCAAG--ACCUUCGCAAUUCAGGAGCCAAAGCCCCUGGC -----------(((....(((((((.(((--------.......((.(((.(((((((....))))))).))).))--.(((.........))).))))))))))..))) ( -32.40, z-score = -1.99, R) >droEre2.scaffold_4770 11293186 89 + 17746568 -----------GCCCCCUGGGCUUUUGGC--------ACACUCACUUGCCAUUAAGUUUUUUGAUUGAGCGGCAGG--ACCUUUGCAAUUCAGGAGCCAAAGCCCUUGGC -----------(((....(((((((.(((--------.......((((((....((((....))))....))))))--.(((.........))).))))))))))..))) ( -31.00, z-score = -1.35, R) >droAna3.scaffold_13340 16375231 97 + 23697760 AACCUGAAGGUGCCCUAAAGCCACGCAGACACA-UAAAUACACAUAC-CCGUUACGGGCCUUGAUUGAGUG---AGAGGACCCAGUGGCA-GGGCCCCAAGGC------- ..(((...((.(((((...(((((...(((...-.............-..)))..(((((((.........---.)))).))).))))))-))))))..))).------- ( -31.47, z-score = -1.73, R) >dp4.chr2 20466321 95 + 30794189 --------UACGGCGUACGGCCUUCAGGGCCUACUGUAUACUCGUU--CCAUUCAGCGCUU-GAUUGAGUG-CGAACGAGCCCUGAGACC---GACCGAAGCCCCAUGGC --------..(((....(((..((((((((...........(((((--((((((((.....-..)))))))-.)))))))))))))).))---).)))..(((....))) ( -34.20, z-score = -2.14, R) >consensus ___________GCCCCCUGGGCUUUUGGCCAUC_UACACACUCACUCGCCAUUCAGUUUUUUGAUUGAGCGGCAGGGGACCAUCGCAAUUCAGGAGCCAAAGCCCUUGGC ...........(((....(((((((.(((..................(((.(((((((....))))))).)))......((...........)).))))))))))..))) (-11.97 = -15.84 + 3.87)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:18:36 2011