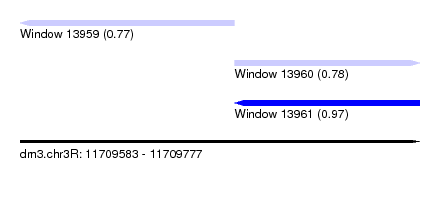
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,709,583 – 11,709,777 |
| Length | 194 |
| Max. P | 0.967305 |

| Location | 11,709,583 – 11,709,687 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 65.67 |
| Shannon entropy | 0.54735 |
| G+C content | 0.59631 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -18.36 |
| Energy contribution | -14.64 |
| Covariance contribution | -3.72 |
| Combinations/Pair | 1.63 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.774399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11709583 104 - 27905053 -----UUUGCUCCGCCCCGCUUGUCGCUAACUCAGGACACAGCACCGCGUUACAGGUU-----UUUUUUUAGAGCGCGACGCCUCCGGAAGUCGGGACAUCGAGUUGGCAGGAG -----....((((((...)).....((((((((........((..((((((..(((..-----....)))..))))))..)).(((.(....).)))....)))))))).)))) ( -31.40, z-score = 0.20, R) >droPer1.super_3 3141011 109 - 7375914 UCCUAUCCUCCCCCCCUACCCUGUCCGCACGGCUGUCGUUAACAUUGUGCU-CAGGUCCACAACUUUCCCAGGGCACACCGUGUCCGGAA----AGACCUCCUGUAGGCGGGGC ..........(((((((((((((...((((((.(((.....))))))))).-)))).......((((((..((((((...))))))))))----)).......))))).)))). ( -42.00, z-score = -3.10, R) >dp4.chr2 20393637 109 - 30794189 UCCUAUCCUCCCCCCCUACCCUGUCCGCACGGCUGUCGUUAACAUUGUGCU-CAGGUCCACAACUUUCCCAGGGCACAUCGUGUCCGGAA----AGACCUCCUGUAGGCGGGGC ..........(((((((((((((...((((((.(((.....))))))))).-)))).......((((((..((((((...))))))))))----)).......))))).)))). ( -42.00, z-score = -3.31, R) >droSim1.chr3R 17752871 103 + 27517382 -----UUCGUUCCG-CCCGCUGGUCGCCAACUCAGGACACAGCAACGCGUUACAGGUU-----UUUUUUUAGAGCGCGACGCCUCCGGAAGUCGGGACAUGGGGUUGGCAGGAG -----.....((((-((....))).((((((((........((..((((((..(((..-----....)))..))))))..)).(((.(....).)))....)))))))).))). ( -32.90, z-score = 0.21, R) >droSec1.super_0 10876005 103 + 21120651 -----UUCGCUCCG-CCCGCUUGUCGCCAACUCAGGACACAGCAACGCGUUACAGGUU-----UUUUUUUAGAGCGCGACGCCUCCGGAAGUCGGGACAUCGGGUUGGCAGGAG -----.((.(((((-((((..((((.((.((((.(((....((..((((((..(((..-----....)))..))))))..)).))).).))).)))))).)))).))).)))). ( -35.20, z-score = -0.71, R) >consensus _____UUCGCUCCGCCCCGCUUGUCGCCAACUCAGGACACAGCAACGCGUUACAGGUU_____UUUUUUUAGAGCGCGACGCCUCCGGAAGUCGGGACAUCGGGUUGGCAGGAG ...................(((((((((......)).)...((..((((((...((...........))...))))))..))........................)))))).. (-18.36 = -14.64 + -3.72)

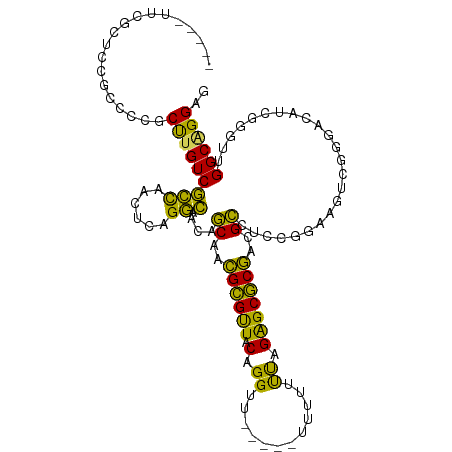
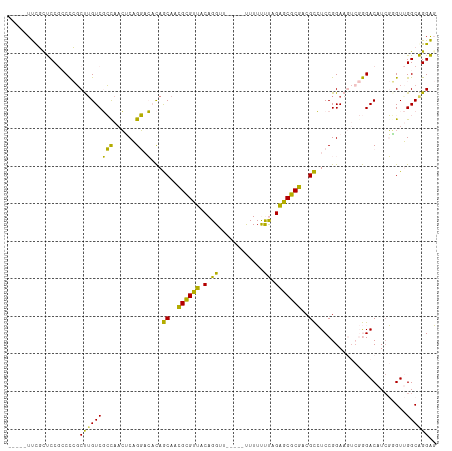
| Location | 11,709,687 – 11,709,777 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 89.68 |
| Shannon entropy | 0.16958 |
| G+C content | 0.55462 |
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -25.78 |
| Energy contribution | -25.78 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.780841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11709687 90 + 27905053 GGGAUG--AUGGCAAGUGCAUAAAGCACCGACAGAUAGCUUCGAUGGGACACCAAGCCAGAACGGAGCCA---------GUGGCUCAGCGCCAUCGAUGAU ....((--(((((..((((.....)))).........(((....(((....)))..........(((((.---------..)))))))))))))))..... ( -29.00, z-score = -1.51, R) >droSim1.chr3R 17752974 90 - 27517382 GGGAUG--AUGGCAAGUGCAUAAAGCACCGACAGAUAGCUUCGAUGGGACACCAAGCCAGGACGGAGCCA---------GUGGCUCAGCGCCAUCGAUGAU ....((--(((((..((((.....)))).........(((....(((....)))..........(((((.---------..)))))))))))))))..... ( -29.00, z-score = -1.25, R) >droSec1.super_0 10876108 90 - 21120651 GGGAUG--AUGGCAAGUGCAUGAAGCACCAAAAGAUAGCUUCGAUGGGACACCAAGCCAGAACGGAGCCA---------GUGGCUCAGCGCCAUCGAUGAU ....((--(((((..(.((.((((((...........)))))).(((....))).)))......(((((.---------..)))))...)))))))..... ( -29.50, z-score = -1.66, R) >droYak2.chr3R 14105079 96 - 28832112 -----GGGAUGGCAAGUGCAUAAAGCACCGACAGAUAGCUUCAAUGGGACACCAAACCAGAACGGAGCCAUCGGAGCCAGUGGCUCAGCGCCAUCGAUGAU -----..((((((..((((.....)))).....(((.(((((..(((.........)))....))))).))).(((((...)))))...))))))...... ( -34.20, z-score = -2.78, R) >droEre2.scaffold_4770 11234814 92 + 17746568 GGGAUGGGAUGGCAAGUGCAUAAAGCACCGACAGAUAGCUUCAAUGGGACACCAAACCAGGACGGAGCCA---------GUGGCUCAGCGCCAUCGAUGAU .......((((((..((((.....)))).........(((....(((....)))..........(((((.---------..))))))))))))))...... ( -28.70, z-score = -1.20, R) >consensus GGGAUG__AUGGCAAGUGCAUAAAGCACCGACAGAUAGCUUCGAUGGGACACCAAGCCAGAACGGAGCCA_________GUGGCUCAGCGCCAUCGAUGAU ........(((((..((((.....)))).........(((....(((....)))..........((((((..........))))))))))))))....... (-25.78 = -25.78 + -0.00)
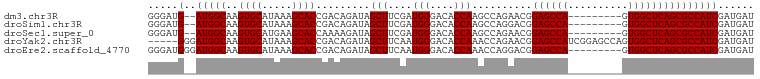
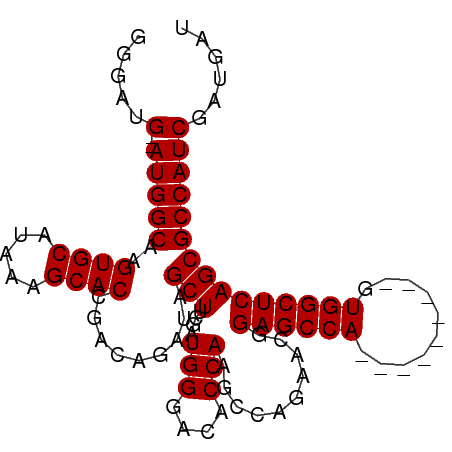
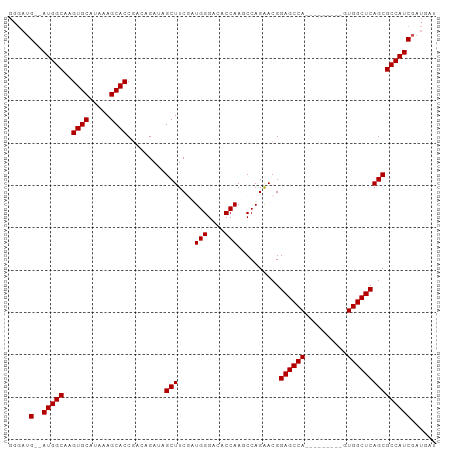
| Location | 11,709,687 – 11,709,777 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 89.68 |
| Shannon entropy | 0.16958 |
| G+C content | 0.55462 |
| Mean single sequence MFE | -32.06 |
| Consensus MFE | -28.22 |
| Energy contribution | -27.94 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11709687 90 - 27905053 AUCAUCGAUGGCGCUGAGCCAC---------UGGCUCCGUUCUGGCUUGGUGUCCCAUCGAAGCUAUCUGUCGGUGCUUUAUGCACUUGCCAU--CAUCCC ....((((((((((..(((((.---------((....))...)))))..)))..)))))))........((.(((((.....))))).))...--...... ( -32.20, z-score = -2.46, R) >droSim1.chr3R 17752974 90 + 27517382 AUCAUCGAUGGCGCUGAGCCAC---------UGGCUCCGUCCUGGCUUGGUGUCCCAUCGAAGCUAUCUGUCGGUGCUUUAUGCACUUGCCAU--CAUCCC ....((((((((((..(((((.---------.(((...))).)))))..)))..)))))))........((.(((((.....))))).))...--...... ( -32.90, z-score = -2.56, R) >droSec1.super_0 10876108 90 + 21120651 AUCAUCGAUGGCGCUGAGCCAC---------UGGCUCCGUUCUGGCUUGGUGUCCCAUCGAAGCUAUCUUUUGGUGCUUCAUGCACUUGCCAU--CAUCCC ......((((((((..(((((.---------((....))...)))))..))....(((.(((((.(((....))))))))))).....)))))--)..... ( -32.00, z-score = -2.65, R) >droYak2.chr3R 14105079 96 + 28832112 AUCAUCGAUGGCGCUGAGCCACUGGCUCCGAUGGCUCCGUUCUGGUUUGGUGUCCCAUUGAAGCUAUCUGUCGGUGCUUUAUGCACUUGCCAUCCC----- ......((((((((((((((...)))))((((((..(((........)))....)))))).)))........(((((.....))))).))))))..----- ( -34.30, z-score = -2.21, R) >droEre2.scaffold_4770 11234814 92 - 17746568 AUCAUCGAUGGCGCUGAGCCAC---------UGGCUCCGUCCUGGUUUGGUGUCCCAUUGAAGCUAUCUGUCGGUGCUUUAUGCACUUGCCAUCCCAUCCC ......(((((.((.(((((..---------.))))).))..((((..(((((.....((((((.(((....))))))))).))))).))))..))))).. ( -28.90, z-score = -1.51, R) >consensus AUCAUCGAUGGCGCUGAGCCAC_________UGGCUCCGUUCUGGCUUGGUGUCCCAUCGAAGCUAUCUGUCGGUGCUUUAUGCACUUGCCAU__CAUCCC ....((((((((((..(((((.....................)))))..)))..)))))))........((.(((((.....))))).))........... (-28.22 = -27.94 + -0.28)
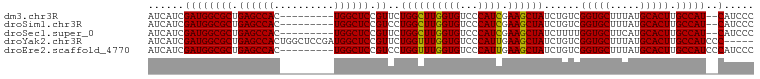
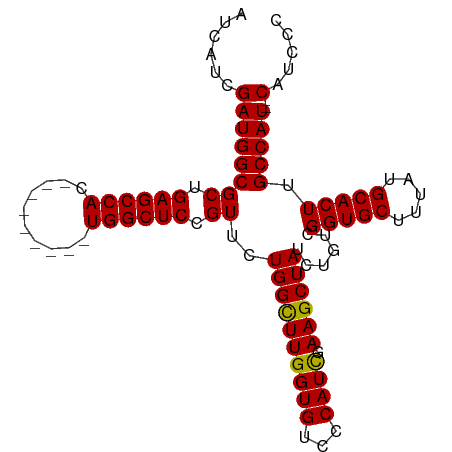
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:18:29 2011