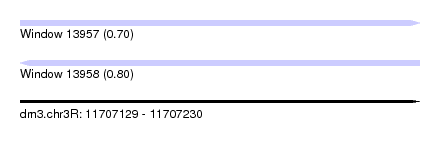
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,707,129 – 11,707,230 |
| Length | 101 |
| Max. P | 0.798560 |

| Location | 11,707,129 – 11,707,230 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 73.24 |
| Shannon entropy | 0.49571 |
| G+C content | 0.47175 |
| Mean single sequence MFE | -34.44 |
| Consensus MFE | -12.47 |
| Energy contribution | -13.80 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.701217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
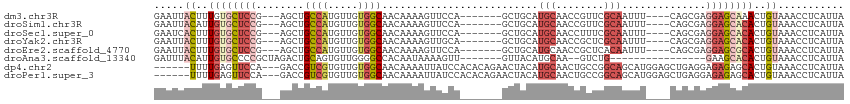
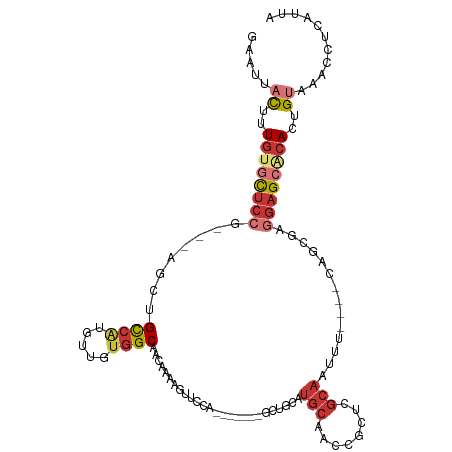
>dm3.chr3R 11707129 101 + 27905053 UAAUGAGGUUUACAGUUUGCUCCUCGCUG----AAAUUGCGAACGGUUGCAUGCAGC-------UGGAACUUUUGUUGCCACAACAUGGCAGCU---CGGAGCACAAAGUAAUUC .........((((..(.((((((((((..----.....)))).((((((....))))-------))........(((((((.....))))))).---.)))))).)..))))... ( -33.70, z-score = -2.21, R) >droSim1.chr3R 17750443 101 - 27517382 UAAUGAGGUUUACAGUGUGCUCCUCGCUG----AAAUUGCGAACGGUUGCAUGCAGC-------UGGAACUUUUGUUGCCACAACAUGGCAGCU---CGGAGCACAAUGUAAUUC .........(((((.((((((((((((..----.....)))).((((((....))))-------))........(((((((.....))))))).---.)))))))).)))))... ( -40.90, z-score = -4.08, R) >droSec1.super_0 10873576 101 - 21120651 UAAUGAGGUUUACAGUGUGCUCCUCGCUG----AAAUUGCGAAAGGUUGCAUGCAGC-------UGGAACUUUUGUUGCCACAACAUGGCAGCU---CGGAGCACAAAGUGAUUC .........((((..((((((((..((((----....(((((....)))))..))))-------..........(((((((.....))))))).---.))))))))..))))... ( -38.30, z-score = -3.05, R) >droYak2.chr3R 14102537 101 - 28832112 UAAUGAGGUUUACAGUGUGCUCCUCGCUG----AAAUUGCGAGCGGUUGCAUGCAGC-------UGCAACUUUUGUUGCCACAACAUGGCAGCU---CGGAGCACAAAGUAAUUC .........((((..((((((((((((..----.....))))(((((((....))))-------))).......(((((((.....))))))).---.))))))))..))))... ( -43.20, z-score = -4.14, R) >droEre2.scaffold_4770 11232388 101 + 17746568 UAAUGAGGUUUACAGUGCGCUCCUCGCUG----AAAUUGUGAGCGGUUGCAUGCAGC-------UGGAACUUUUGUUGCCACAACAUGGCAGCU---CGGAGCACAAAGUAAUUC .........((((..((.(((((((((..----.....)))).((((((....))))-------))........(((((((.....))))))).---.))))).))..))))... ( -33.60, z-score = -1.34, R) >droAna3.scaffold_13340 16312448 90 + 23697760 UAAUGAGGUUUACAGUGUGCUUC----------------CAGAC--UUGCAUGUAAC-------AACUUUUAUUGUGGCCCCAACACUGCAGUCUAGCGGGGCACAAUGUAAAUC ......((((((((.((((((((----------------(((((--(.((((((.((-------((......))))(....).))).)))))))).).)))))))).)))))))) ( -30.00, z-score = -2.55, R) >dp4.chr2 20391506 106 + 30794189 UAAUGAGGUUUACAGUGCUCUCUCCUCAGCUCCAUGCUGCCGGCAGUUGCAUGUAGUUCUGUGUGGAUAAUUUUGUUGCCACAACACGACGGUC---UGGAACUCAAAA------ ...((((...((((.(((...((((.((((.....))))..)).))..)))))))(((.(((((((.((((...)))))))).))).)))....---.....))))...------ ( -27.90, z-score = -0.06, R) >droPer1.super_3 3138857 106 + 7375914 UAAUGAGGUUUACAGUGCUCUCUCCUCAGCUCCAUGCUGCCGGCAGUUGCAUGUAGUUCUGUGUGGAUAAUUUUGUUGCCACAACACGACGGUC---UGGAACUCAAAA------ ...((((...((((.(((...((((.((((.....))))..)).))..)))))))(((.(((((((.((((...)))))))).))).)))....---.....))))...------ ( -27.90, z-score = -0.06, R) >consensus UAAUGAGGUUUACAGUGUGCUCCUCGCUG____AAAUUGCGAACGGUUGCAUGCAGC_______UGGAACUUUUGUUGCCACAACAUGGCAGCU___CGGAGCACAAAGUAAUUC .........((((..((.(((((..............(((((....))))).......................((((((.......)))))).....))))).))..))))... (-12.47 = -13.80 + 1.33)

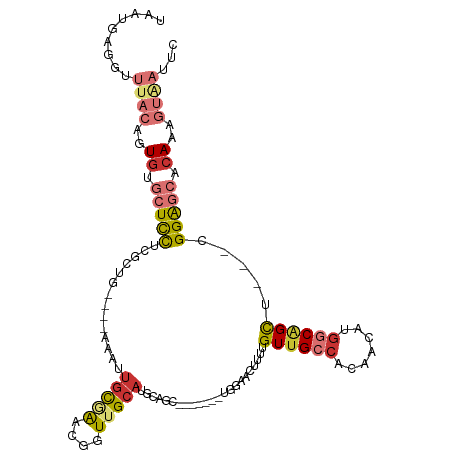
| Location | 11,707,129 – 11,707,230 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 73.24 |
| Shannon entropy | 0.49571 |
| G+C content | 0.47175 |
| Mean single sequence MFE | -31.36 |
| Consensus MFE | -13.26 |
| Energy contribution | -14.46 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.798560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11707129 101 - 27905053 GAAUUACUUUGUGCUCCG---AGCUGCCAUGUUGUGGCAACAAAAGUUCCA-------GCUGCAUGCAACCGUUCGCAAUUU----CAGCGAGGAGCAAACUGUAAACCUCAUUA ...((((....((((((.---.(((((...((((..((.......))..))-------)).))).))......((((.....----..))))))))))....))))......... ( -30.00, z-score = -1.78, R) >droSim1.chr3R 17750443 101 + 27517382 GAAUUACAUUGUGCUCCG---AGCUGCCAUGUUGUGGCAACAAAAGUUCCA-------GCUGCAUGCAACCGUUCGCAAUUU----CAGCGAGGAGCACACUGUAAACCUCAUUA ...(((((.((((((((.---.(((((...((((..((.......))..))-------)).))).))......((((.....----..)))))))))))).)))))......... ( -35.90, z-score = -3.51, R) >droSec1.super_0 10873576 101 + 21120651 GAAUCACUUUGUGCUCCG---AGCUGCCAUGUUGUGGCAACAAAAGUUCCA-------GCUGCAUGCAACCUUUCGCAAUUU----CAGCGAGGAGCACACUGUAAACCUCAUUA .....((..((((((((.---.(((((...((((..((.......))..))-------)).))).))......((((.....----..))))))))))))..))........... ( -32.30, z-score = -2.50, R) >droYak2.chr3R 14102537 101 + 28832112 GAAUUACUUUGUGCUCCG---AGCUGCCAUGUUGUGGCAACAAAAGUUGCA-------GCUGCAUGCAACCGCUCGCAAUUU----CAGCGAGGAGCACACUGUAAACCUCAUUA ...((((..(((((((((---(((((((((...))))))......((((((-------......)))))).))))((.....----..))..))))))))..))))......... ( -40.70, z-score = -4.19, R) >droEre2.scaffold_4770 11232388 101 - 17746568 GAAUUACUUUGUGCUCCG---AGCUGCCAUGUUGUGGCAACAAAAGUUCCA-------GCUGCAUGCAACCGCUCACAAUUU----CAGCGAGGAGCGCACUGUAAACCUCAUUA ...((((..((((((((.---.((((....(((((((((((....)))...-------((.....))....)).))))))..----))))..))))))))..))))......... ( -36.20, z-score = -3.42, R) >droAna3.scaffold_13340 16312448 90 - 23697760 GAUUUACAUUGUGCCCCGCUAGACUGCAGUGUUGGGGCCACAAUAAAAGUU-------GUUACAUGCAA--GUCUG----------------GAAGCACACUGUAAACCUCAUUA ..((((((.(((((....((((((((((.(((((......)))))...((.-------...)).))).)--)))))----------------)..))))).))))))........ ( -27.80, z-score = -1.89, R) >dp4.chr2 20391506 106 - 30794189 ------UUUUGAGUUCCA---GACCGUCGUGUUGUGGCAACAAAAUUAUCCACACAGAACUACAUGCAACUGCCGGCAGCAUGGAGCUGAGGAGAGAGCACUGUAAACCUCAUUA ------.....(((((((---..(.(((((((.((((............))))))).........((....)))))).)..)))))))(((((.((....)).)...)))).... ( -24.00, z-score = 0.61, R) >droPer1.super_3 3138857 106 - 7375914 ------UUUUGAGUUCCA---GACCGUCGUGUUGUGGCAACAAAAUUAUCCACACAGAACUACAUGCAACUGCCGGCAGCAUGGAGCUGAGGAGAGAGCACUGUAAACCUCAUUA ------.....(((((((---..(.(((((((.((((............))))))).........((....)))))).)..)))))))(((((.((....)).)...)))).... ( -24.00, z-score = 0.61, R) >consensus GAAUUACUUUGUGCUCCG___AGCUGCCAUGUUGUGGCAACAAAAGUUCCA_______GCUGCAUGCAACCGCUCGCAAUUU____CAGCGAGGAGCACACUGUAAACCUCAUUA .....((..((((((((........((((.....))))..........................(((........)))..............))))))))..))........... (-13.26 = -14.46 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:18:27 2011