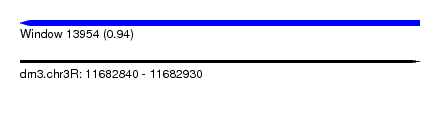
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,682,840 – 11,682,930 |
| Length | 90 |
| Max. P | 0.940042 |

| Location | 11,682,840 – 11,682,930 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 58.36 |
| Shannon entropy | 0.74996 |
| G+C content | 0.43769 |
| Mean single sequence MFE | -22.94 |
| Consensus MFE | -8.15 |
| Energy contribution | -7.93 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.940042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11682840 90 - 27905053 AUCUCUUGGUGCCGUGCUGCAA-----GGGCACCAA-----------UUAUCAAUU------CACAAUCUGCACUCCGCUUACCCGGAGG-AAAAAAAUA-GUGAUUUCCCCUU .....((((((((.(......)-----.))))))))-----------........(------(((........(((((......))))).-.........-))))......... ( -23.97, z-score = -1.90, R) >droSim1.chr3R 17726576 78 + 27517382 AUCUUUUGGUGCCGUGCUGCAA-----AGGCACCAA-----------UUAUCAAUU------CACUAUUUGCACUCCGC-------------AAAAAAGA-AUGAUUUUCCCUU .....((((((((.........-----.))))))))-----------..((((.((------(....(((((.....))-------------)))...))-)))))........ ( -19.10, z-score = -2.60, R) >droSec1.super_0 10848546 78 + 21120651 AUCUCUUGGUGCCGUGCUGCAA-----AGGCACCAA-----------UUAUCAAUU------CACUAUUUGCACUCCGC-------------AAAAAAGA-AUGAUUUUCCCUU .....((((((((.........-----.))))))))-----------..((((.((------(....(((((.....))-------------)))...))-)))))........ ( -19.00, z-score = -2.71, R) >droYak2.chr3R 14077421 102 + 28832112 GUCUCCUGGUGCCGUGCUGAAA-----AGUCACCAAAUUACAUAUACUUAUCAAAU------CACAAUUUGCACUCCGCUUACCUGGUGGGCAAAAAAUA-GUGAUUUCCCUUU ......(((((....(((....-----)))))))).................((((------(((.((((....((((((.....))))))....)))).-)))))))...... ( -22.60, z-score = -1.77, R) >droEre2.scaffold_4770 11207487 99 - 17746568 ACCUCCUGGUGCCGUGCUGAAA-----AACCACUACAUUAAAUAUACUUAUCAAAU------CACAAUUUGCACUCCGCUUACCUGGAGGCUAAAA---A-GUGAUUUCCCUUU (((....)))...(((.((...-----...)).)))................((((------(((..(((((.(((((......))))))).))).---.-)))))))...... ( -18.30, z-score = -1.74, R) >dp4.chr2 20361278 114 - 30794189 GAACUUUGGUGCCGUGCCAGAAAGAGUAGGUGCUAAUAGUAGAACAUUCGACACCCUUUUUGCGCACCUAUCUCUUCCCGUGUGGUGUGGUCGAGGCAAAUGUGAUUCCCGUUU ...(((((..((((..(((.(((((((((((((.....((((((.............))))))))))))).)))))....).)))..))))))))).................. ( -30.92, z-score = -0.63, R) >droPer1.super_3 3112876 114 - 7375914 GAACUUUGGUGCCGUGCCAGAAAGAGUAGGUGCUAAUAGUAGAACAUUCGACACCCUUUUUGCGCACCUAUCUCUUCCCGUGUGAUGUCGUCGAGGCAAAUGUGAUUCCCGUUU ((((((((((.....))))))((((((((((((.....((((((.............))))))))))))).)))))..((..(..((((.....)))).)..))......)))) ( -26.72, z-score = -0.09, R) >consensus AUCUCUUGGUGCCGUGCUGCAA_____AGGCACCAA_______A_A_UUAUCAAUU______CACAAUUUGCACUCCGCUUACC_GGUGG_CAAAAAAAA_GUGAUUUCCCUUU ......(((((((...............)))))))............................................................................... ( -8.15 = -7.93 + -0.22)

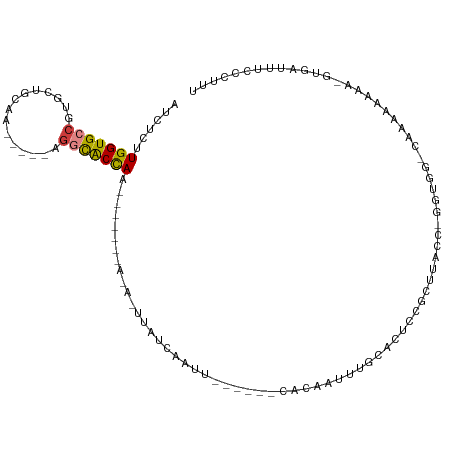
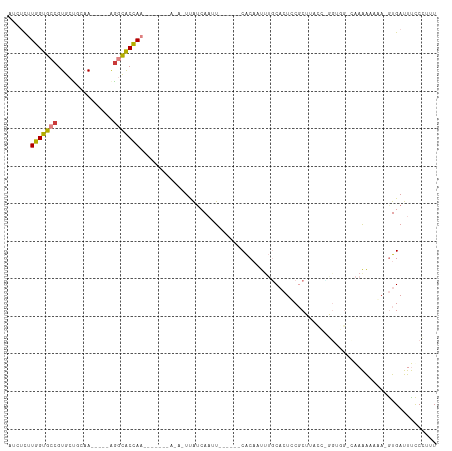
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:18:24 2011