
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,671,037 – 11,671,157 |
| Length | 120 |
| Max. P | 0.736596 |

| Location | 11,671,037 – 11,671,155 |
|---|---|
| Length | 118 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.64 |
| Shannon entropy | 0.59281 |
| G+C content | 0.50092 |
| Mean single sequence MFE | -37.48 |
| Consensus MFE | -14.14 |
| Energy contribution | -13.89 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.707031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
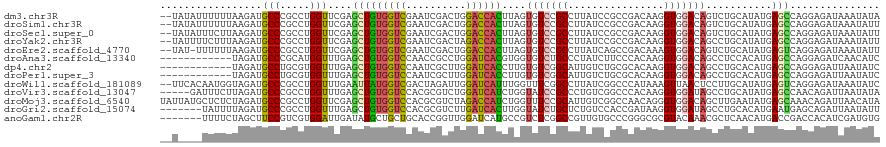
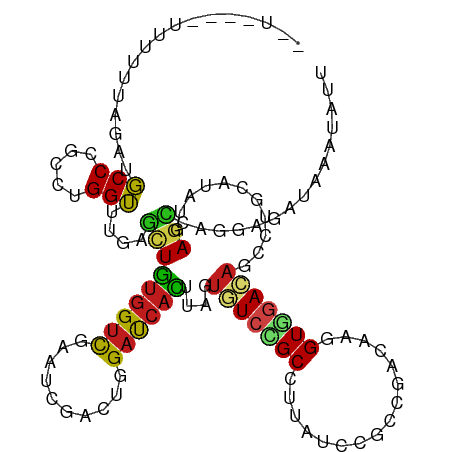
>dm3.chr3R 11671037 118 - 27905053 --UAUAUUUUUUAAGAUGCCCGCCUGGUUCGAGCUGUGGUCGAAUCGACUGGACCACUUAGUGUCCGCCUUAUCCGCCGACAAGGUGGACAGUCUGCAUAUGAGCCAGGAGAUAAAUAUA --..................(.(((((((((.((.((((((.(......).))))))....((((((((((.((....)).))))))))))....))...))))))))).)......... ( -42.00, z-score = -3.38, R) >droSim1.chr3R 17714112 118 + 27517382 --UAUAUUUUUUAAGAUGCCCGCCUGGUUCGAGCUGUGGUCGAAUCGACUGGACCACUUAGUGUCCGCCUUAUCCGCCGACAAGGUGGACAGUCUGCAUAUGAGCCAGGAGAUAAAUAUU --..................(.(((((((((.((.((((((.(......).))))))....((((((((((.((....)).))))))))))....))...))))))))).)......... ( -42.00, z-score = -3.44, R) >droSec1.super_0 10836810 118 + 21120651 --UAUAUUUCUUAAGAUGCCCGCCUGGUUCGAGCUGUGGUCGAAUCGACUGGACCACUUAGUGUCCGCCUUAUCCGCCGACAAGGUGGACAGUCUGCAUAUGAGCCAGGAGAUAAAUAUU --..................(.(((((((((.((.((((((.(......).))))))....((((((((((.((....)).))))))))))....))...))))))))).)......... ( -42.00, z-score = -3.18, R) >droYak2.chr3R 14065624 118 + 28832112 --UAUUUUCUUUAAGAUGCCCGCCUGGUUCGAGCUGUGGUCGAAUCGACUAGACCACUUAGUGUCCGCCUUAUCCGCCGACAAGGUGGACAGCCUGCAUAUGAGCCAGGAGAUAAAUAUU --..................(.(((((((((.((.((((((..........))))))....((((((((((.((....)).))))))))))....))...))))))))).)......... ( -41.60, z-score = -3.59, R) >droEre2.scaffold_4770 11195753 117 - 17746568 --UAU-UUUUUUAAGAUGCCCGCCUGGUUCGAGCUGUGGUCGAAUCGACUGGACCACUUAGUGUCCGCCUUAUCAGCCGACAAAGUGGACAGUCUGCAUAUGAGUCAGGAGAUAAAUAUU --(((-(((((....((((......(((((((.(...).)))))))(((((..((((((..((((.((.......)).)))))))))).))))).)))).......))))))))...... ( -34.30, z-score = -1.65, R) >droAna3.scaffold_13340 16276020 108 - 23697760 ------------UAGAUGCCCGCAUGGUUUGAGCUGUGGUCCAACCGCCUGGAUCACGUGGUGUCUGCCCUAUCUUCCCACAAGGUGGACAGCCUCCACAUGAGCCAGGAGAUCAACAUC ------------..(((..((...((((((.....((((((((......))))))))((((.(.(((..(((((((.....))))))).))).).))))..))))))))..)))...... ( -36.70, z-score = -0.83, R) >dp4.chr2 20349706 108 - 30794189 ------------UAGAUGCCUGCGUGGUUUGAGCUGUGGUCCAAUCGCUUGGAUCACCUUGUGUCGGCAUUGUCUGCGCACAAGGUGGACAGCCUGCACAUGAGCCAGGAGAUUAAUAUC ------------......((((((((((..(.((((((((((((....)))))))(((((((((..(((.....))))))))))))..)))))).)).))))...))))........... ( -38.60, z-score = -1.10, R) >droPer1.super_3 3101256 108 - 7375914 ------------UAGAUGCCUGCGUGGUUUGAGCUGUGGUCCAAUCGCUUGGAUCACCUUGUGUCGGCAUUGUCUGCGCACAAGGUGGACAGCCUGCACAUGAGCCAGGAGAUUAAUAUC ------------......((((((((((..(.((((((((((((....)))))))(((((((((..(((.....))))))))))))..)))))).)).))))...))))........... ( -38.60, z-score = -1.10, R) >droWil1.scaffold_181089 7181620 118 + 12369635 --UUCACAAUGGUAGAUGCCCGCCUGGUUUGAAUUAUGGUCGACUAGAUUGGAUCAUUUGGUUUCGGCCUUAUCGGCCCAUAAAGUUAACUCCUUGCAUAUGAGUCAGGAGAUAAAUAUC --......(((...((.(((....(((((..(....(((....)))..)..)))))...))).))((((.....)))))))........(((((.((......)).)))))......... ( -26.30, z-score = 0.65, R) >droVir3.scaffold_13047 2321603 115 - 19223366 -----GAUUUCUUAGAUGCCCGCCUGGUUUGAGCUGUGGUCCACGCGUCUGGAUCAUCUGGUAUCCGCCCUGUCGGCCCACAAGGUGGAUAGCCUGCAUAUGAGCCAACAGAUUAAUAUA -----((((...(((((((..(((((((....)))).)))....)))))))))))(((((.((((((((.(((......))).))))))))....((......))...)))))....... ( -32.50, z-score = 0.37, R) >droMoj3.scaffold_6540 18805378 120 + 34148556 UAUUAUGCUCUCUAGAUGCCCGCCUGGUUCGAGCUGUGGUCCACGCGUCUAGACCAUCUGGUUUCCGCAUUGUCGGCCAACAGGGUGGACAGCUUGAAUAUGAGCAAACAGAUUAACAUA .....(((((.((((........))))((((((((...(((((((((...(((((....))))).)))........((....))))))))))))))))...))))).............. ( -37.20, z-score = -0.93, R) >droGri2.scaffold_15074 5891176 113 - 7742996 -------UAUUUUAGAUGCCCGCCUGGUUUGAGCUGUGGUCCACGCGUCUUGAUCACUUGGUAUCUGCUCUGUCCACCGAUAAGGUGGAUAGCCUGCACAUGAAUGAGCAGAUUAAUAUU -------.....((((((((....(((.(..((.((((.....)))).))..))))...))))))))..(((((((((.....))))))))).((((.((....)).))))......... ( -37.70, z-score = -2.48, R) >anoGam1.chr2R 14099477 113 - 62725911 -------UUUUCUAGCUUCCGUCGUGGAUUGAUAUGCUGCUGCACCGGUUGGAUCAUGCCGUCUCGGCCGUUGUGCCCGGGCGCGUACAAACGCUCAACAUGACCGACCACAUCGAUGUG -------............(((((((........(((....)))..((((((.(((((..((((.(((......))).))))((((....))))....))))))))))).)).))))).. ( -37.80, z-score = -0.66, R) >consensus __U____UUUUUUAGAUGCCCGCCUGGUUUGAGCUGUGGUCGAAUCGACUGGAUCACUUAGUGUCCGCCUUAUCCGCCGACAAGGUGGACAGCCUGCAUAUGAGCCAGGAGAUAAAUAUU .................(((.....)))....(((((((((..........))))))....(((((((................)))))))...........)))............... (-14.14 = -13.89 + -0.25)
| Location | 11,671,039 – 11,671,157 |
|---|---|
| Length | 118 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.22 |
| Shannon entropy | 0.59896 |
| G+C content | 0.50165 |
| Mean single sequence MFE | -37.63 |
| Consensus MFE | -14.14 |
| Energy contribution | -13.89 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.736596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
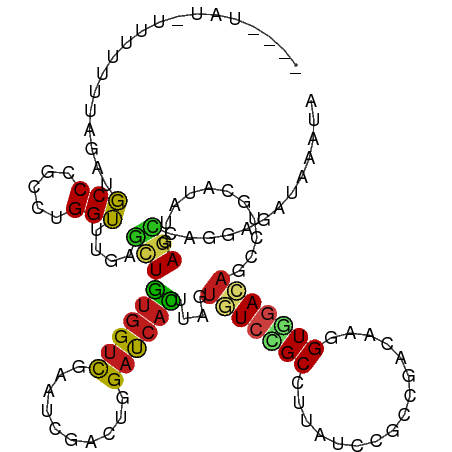
>dm3.chr3R 11671039 118 - 27905053 --UUUAUAUUUUUUAAGAUGCCCGCCUGGUUCGAGCUGUGGUCGAAUCGACUGGACCACUUAGUGUCCGCCUUAUCCGCCGACAAGGUGGACAGUCUGCAUAUGAGCCAGGAGAUAAAUA --....................(.(((((((((.((.((((((.(......).))))))....((((((((((.((....)).))))))))))....))...))))))))).)....... ( -42.00, z-score = -3.42, R) >droSim1.chr3R 17714114 118 + 27517382 --UUUAUAUUUUUUAAGAUGCCCGCCUGGUUCGAGCUGUGGUCGAAUCGACUGGACCACUUAGUGUCCGCCUUAUCCGCCGACAAGGUGGACAGUCUGCAUAUGAGCCAGGAGAUAAAUA --....................(.(((((((((.((.((((((.(......).))))))....((((((((((.((....)).))))))))))....))...))))))))).)....... ( -42.00, z-score = -3.42, R) >droSec1.super_0 10836812 118 + 21120651 --UUUAUAUUUCUUAAGAUGCCCGCCUGGUUCGAGCUGUGGUCGAAUCGACUGGACCACUUAGUGUCCGCCUUAUCCGCCGACAAGGUGGACAGUCUGCAUAUGAGCCAGGAGAUAAAUA --....................(.(((((((((.((.((((((.(......).))))))....((((((((((.((....)).))))))))))....))...))))))))).)....... ( -42.00, z-score = -3.16, R) >droYak2.chr3R 14065626 118 + 28832112 --UUUAUUUUCUUUAAGAUGCCCGCCUGGUUCGAGCUGUGGUCGAAUCGACUAGACCACUUAGUGUCCGCCUUAUCCGCCGACAAGGUGGACAGCCUGCAUAUGAGCCAGGAGAUAAAUA --(((((((((.....))......(((((((((.((.((((((..........))))))....((((((((((.((....)).))))))))))....))...)))))))))))))))).. ( -41.80, z-score = -3.60, R) >droEre2.scaffold_4770 11195755 117 - 17746568 --UUUAU-UUUUUUAAGAUGCCCGCCUGGUUCGAGCUGUGGUCGAAUCGACUGGACCACUUAGUGUCCGCCUUAUCAGCCGACAAAGUGGACAGUCUGCAUAUGAGUCAGGAGAUAAAUA --(((((-(((((....((((......(((((((.(...).)))))))(((((..((((((..((((.((.......)).)))))))))).))))).)))).......)))))))))).. ( -35.90, z-score = -2.08, R) >droAna3.scaffold_13340 16276022 108 - 23697760 ------------CUUAGAUGCCCGCAUGGUUUGAGCUGUGGUCCAACCGCCUGGAUCACGUGGUGUCUGCCCUAUCUUCCCACAAGGUGGACAGCCUCCACAUGAGCCAGGAGAUCAACA ------------....(((..((...((((((.....((((((((......))))))))((((.(.(((..(((((((.....))))))).))).).))))..))))))))..))).... ( -36.70, z-score = -0.74, R) >dp4.chr2 20349708 108 - 30794189 ------------GAUAGAUGCCUGCGUGGUUUGAGCUGUGGUCCAAUCGCUUGGAUCACCUUGUGUCGGCAUUGUCUGCGCACAAGGUGGACAGCCUGCACAUGAGCCAGGAGAUUAAUA ------------........((((((((((..(.((((((((((((....)))))))(((((((((..(((.....))))))))))))..)))))).)).))))...))))......... ( -38.60, z-score = -0.97, R) >droPer1.super_3 3101258 108 - 7375914 ------------GAUAGAUGCCUGCGUGGUUUGAGCUGUGGUCCAAUCGCUUGGAUCACCUUGUGUCGGCAUUGUCUGCGCACAAGGUGGACAGCCUGCACAUGAGCCAGGAGAUUAAUA ------------........((((((((((..(.((((((((((((....)))))))(((((((((..(((.....))))))))))))..)))))).)).))))...))))......... ( -38.60, z-score = -0.97, R) >droWil1.scaffold_181089 7181622 118 + 12369635 --CAUUCACAAUGGUAGAUGCCCGCCUGGUUUGAAUUAUGGUCGACUAGAUUGGAUCAUUUGGUUUCGGCCUUAUCGGCCCAUAAAGUUAACUCCUUGCAUAUGAGUCAGGAGAUAAAUA --..((((.(..((..(....)..))...).))))(((((...((((((((......))))))))..((((.....)))))))))......(((((.((......)).)))))....... ( -26.50, z-score = 0.68, R) >droVir3.scaffold_13047 2321605 115 - 19223366 -----AUGAUUUCUUAGAUGCCCGCCUGGUUUGAGCUGUGGUCCACGCGUCUGGAUCAUCUGGUAUCCGCCCUGUCGGCCCACAAGGUGGAUAGCCUGCAUAUGAGCCAACAGAUUAAUA -----.(((((...(((((((..(((((((....)))).)))....))))))))))))((((.((((((((.(((......))).))))))))....((......))...))))...... ( -32.70, z-score = 0.40, R) >droMoj3.scaffold_6540 18805380 120 + 34148556 GUUAUUAUGCUCUCUAGAUGCCCGCCUGGUUCGAGCUGUGGUCCACGCGUCUAGACCAUCUGGUUUCCGCAUUGUCGGCCAACAGGGUGGACAGCUUGAAUAUGAGCAAACAGAUUAACA ((((...(((((.((((........))))((((((((...(((((((((...(((((....))))).)))........((....))))))))))))))))...))))).......)))). ( -37.50, z-score = -0.83, R) >droGri2.scaffold_15074 5891178 113 - 7742996 -------UUUAUUUUAGAUGCCCGCCUGGUUUGAGCUGUGGUCCACGCGUCUUGAUCACUUGGUAUCUGCUCUGUCCACCGAUAAGGUGGAUAGCCUGCACAUGAAUGAGCAGAUUAAUA -------.......((((((((....(((.(..((.((((.....)))).))..))))...))))))))..(((((((((.....))))))))).((((.((....)).))))....... ( -37.70, z-score = -2.53, R) >anoGam1.chr2R 14099479 111 - 62725911 ---------UUUUCUAGCUUCCGUCGUGGAUUGAUAUGCUGCUGCACCGGUUGGAUCAUGCCGUCUCGGCCGUUGUGCCCGGGCGCGUACAAACGCUCAACAUGACCGACCACAUCGAUG ---------............(((((((........(((....)))..((((((.(((((..((((.(((......))).))))((((....))))....))))))))))).)).))))) ( -37.20, z-score = -0.83, R) >consensus ____UAU_UUUUUUUAGAUGCCCGCCUGGUUUGAGCUGUGGUCGAAUCGACUGGAUCACUUAGUGUCCGCCUUAUCCGCCGACAAGGUGGACAGCCUGCAUAUGAGCCAGGAGAUAAAUA ...................(((.....)))....(((((((((..........))))))....(((((((................)))))))...........)))............. (-14.14 = -13.89 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:18:23 2011