| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,590,629 – 11,590,722 |
| Length | 93 |
| Max. P | 0.993442 |

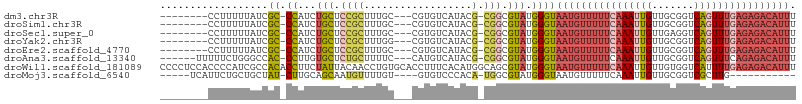
| Location | 11,590,629 – 11,590,722 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 77.75 |
| Shannon entropy | 0.43183 |
| G+C content | 0.45456 |
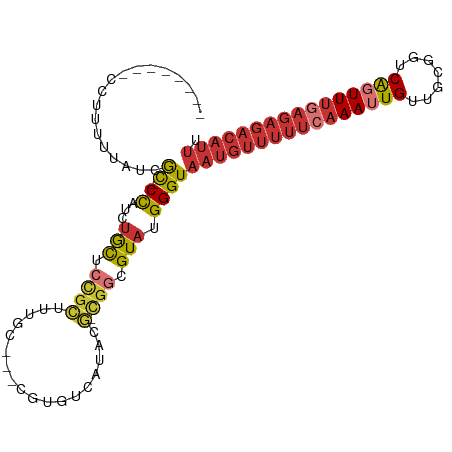
| Mean single sequence MFE | -29.53 |
| Consensus MFE | -15.04 |
| Energy contribution | -16.95 |
| Covariance contribution | 1.91 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.993442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11590629 93 + 27905053 --------CCUUUUUAUCGC-CCAUCUGCUCCGCUUUGC---CGUGUCAUACG-CGGCGUAUGGGUAAUGUUUUUCAAAUUGUUGCGGUCAGUUUGAGAGACAUUU --------..........((-(((((.((...))...((---((((.....))-))))).))))))((((((((((((((((.......)))))))))))))))). ( -34.10, z-score = -4.44, R) >droSim1.chr3R 17622304 93 - 27517382 --------CCUUUUUAUCGC-CCAUCUGCUCCGCUUUGC---CGUGUCAUACG-CGGCGUAUGGGUAAUGUUUUUCAAAUUGUUGCGGUCAGUUUGAGAGACAUUU --------..........((-(((((.((...))...((---((((.....))-))))).))))))((((((((((((((((.......)))))))))))))))). ( -34.10, z-score = -4.44, R) >droSec1.super_0 10756966 93 - 21120651 --------CCUUUUUAUCGC-CCAUCUGCUCCGCUUUGC---CGUGUCAUACG-CGGCGUAUGGGUAAUGUUUUUCAAAUUGUUGAGGUCAGUUUGAGAGACAUUU --------..........((-(((((.((...))...((---((((.....))-))))).))))))((((((((((((((((.......)))))))))))))))). ( -34.10, z-score = -4.62, R) >droYak2.chr3R 13984366 93 - 28832112 --------CCUUUUUAUCGC-CCAUCUGCUCCGCUUUGG---CGUGUCAUACG-CGGCGUAUGGGUAAUGUUUUUCAAAUUGUUGCGGUCAGUUUGAGAGACAUUU --------..........((-((((..((.((((..(((---....)))...)-))).))))))))((((((((((((((((.......)))))))))))))))). ( -33.70, z-score = -3.93, R) >droEre2.scaffold_4770 11101369 93 + 17746568 --------CCUUUUUAUCGC-CCAUCUGCUCCGCUUUGC---CGUGUCAUACG-CGGCGUAUGGGUAAUGUUUUUCAAAUUGUUGCGGUCAGUUUGAGAGACAUUU --------..........((-(((((.((...))...((---((((.....))-))))).))))))((((((((((((((((.......)))))))))))))))). ( -34.10, z-score = -4.44, R) >droAna3.scaffold_13340 16193173 95 + 23697760 ------UUUUUCUGGGCCAC-CCUUGUGCUCUGCUUUUC---CAUGUCAUACG-CGGCGUAUGGGUAAUGUUUUUCAAAUUGUUGCGGUCAGUUUCAGAGACAUUU ------............((-((..((((.((((.....---..........)-))).))))))))(((((((((.((((((.......)))))).))))))))). ( -21.96, z-score = -0.19, R) >droWil1.scaffold_181089 10049662 106 - 12369635 CCCCUCCACCCCAUCGCCACACCUUCUAUUACAACCUGUGCACCUUUCACAUGGCAGCGUAUGGGUAAUGUUUUUCAAAUUGUUGUGGUCAUUUUGAGAGACAUUU .........((((((((((((...............))))..((........))..))).))))).(((((((((((((.((.......)).))))))))))))). ( -25.16, z-score = -1.52, R) >droMoj3.scaffold_6540 22515902 84 + 34148556 -----UCAUUCUGCUGCUAU-CUUGCAGCAAUGUUUUGU----GUGUCCCACA-UGGCGUAUGGGUAAUGUUUUUCAAAUUGUUGCGGUCGCUUG----------- -----((((..((((((...-...))))))((((..(((----(.....))))-..))))))))((((((..........)))))).........----------- ( -19.00, z-score = -0.13, R) >consensus ________CCUUUUUAUCGC_CCAUCUGCUCCGCUUUGC___CGUGUCAUACG_CGGCGUAUGGGUAAUGUUUUUCAAAUUGUUGCGGUCAGUUUGAGAGACAUUU ..............................................((((((......))))))..((((((((((((((((.......)))))))))))))))). (-15.04 = -16.95 + 1.91)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:18:19 2011