
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,585,590 – 11,585,710 |
| Length | 120 |
| Max. P | 0.831683 |

| Location | 11,585,590 – 11,585,686 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.77 |
| Shannon entropy | 0.35686 |
| G+C content | 0.36954 |
| Mean single sequence MFE | -24.24 |
| Consensus MFE | -18.00 |
| Energy contribution | -17.54 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.831683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
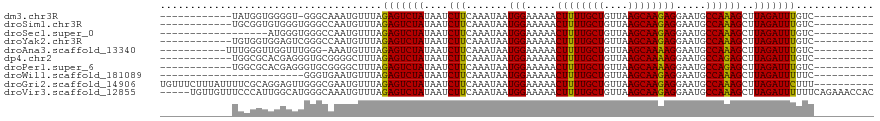
>dm3.chr3R 11585590 96 + 27905053 ------------UAUGGUGGGGU-GGGCAAAUGUUUAGAGUCUAUAAUCUUCAAAUAAUGGAAAAACUUUUGCUGUUAAGCAAGAGGAAUGCCAAAGCUUAGAUUUGUC---------- ------------...((..((.(-((((...(((((..((........))..))))).(((.....((((((((....)))))))).....)))..)))))..))..))---------- ( -23.30, z-score = -1.80, R) >droSim1.chr3R 17617388 97 - 27517382 ------------UGCGGUGUGGGUGGGCCAAUGUUUAGAGUCUAUAAUCUUCAAAUAAUGGAAAAACUUUUGCUGUUAAGCAAGAGGAAUGCCAAAGCUUAGAUUUGUC---------- ------------...(((.(((((.(((((.(((((..((........))..))))).))......((((((((....))))))))....)))...))))).)))....---------- ( -24.30, z-score = -1.34, R) >droSec1.super_0 10752089 91 - 21120651 ------------------AUGGGUGGGCCAAUGUUUAGAGUCUAUAAUCUUCAAAUAAUGGAAAAACUUUUGCUGUUAAGCAAGAGGAAUGCCAAAGCUUAGAUUUGUC---------- ------------------.(((((.(((((.(((((..((........))..))))).))......((((((((....))))))))....)))...)))))........---------- ( -22.70, z-score = -1.69, R) >droYak2.chr3R 13979157 97 - 28832112 ------------UGUGGUGGAGUCGGGCCAAUGUUUAGAGUCUAUAAUCUUCAAAUAAUGGAAAAACUUUUGCUGUUAAGCAAGAGGAAUGCCAAAGCUUAGAUUUGUC---------- ------------.((..(((..((...(((.(((((..((........))..))))).))).....((((((((....))))))))))...)))..))...........---------- ( -24.60, z-score = -1.77, R) >droAna3.scaffold_13340 16187479 97 + 23697760 -----------UUUGGGUUGGUUUGGG-AAAUGUUUAGAGUCUAUAAUCUUCAAAUAAUGGAAAAACUUUUGCUGUUAAGCAAAAGGAAUGCCAAAGCUUAGAUUUGUC---------- -----------((((((((........-...(((((..((........))..))))).(((.....((((((((....)))))))).....))).))))))))......---------- ( -21.30, z-score = -1.47, R) >dp4.chr2 30629896 97 + 30794189 ------------UGGCGCACGAGGGUGCGGGGCUUUAGAGUCUAUAAUCUUCAAAUAAUGGAAAAACUUUUGCUGUUAAGCAAAAGGAAUGCCAGAGCUUAGAUUUGUC---------- ------------.(((.(..((((((...(((((....)))))...))))))......(((.....((((((((....)))))))).....)))).)))..........---------- ( -28.50, z-score = -2.15, R) >droPer1.super_6 6031608 97 + 6141320 ------------UGGCGCACGAGGGUGCGGGGCUUUAGAGUCUAUAAUCUUCAAAUAAUGGAAAAACUUUUGCUGUUAAGCAAAAGGAAUGCCAGAGCUUAGAUUUGUC---------- ------------.(((.(..((((((...(((((....)))))...))))))......(((.....((((((((....)))))))).....)))).)))..........---------- ( -28.50, z-score = -2.15, R) >droWil1.scaffold_181089 10044796 85 - 12369635 ------------------------GGGUGAAUGUUUAGAGUCUAUAAUCUUCAAAUAAUGGAAAAACUUUUGCUGUUAAGCAAGAGGAAUGCCAAAGCUUAGAUUUUUC---------- ------------------------((((...(((((..((........))..))))).(((.....((((((((....)))))))).....)))..)))).........---------- ( -18.00, z-score = -1.14, R) >droGri2.scaffold_14906 4902444 109 - 14172833 UGUUUCUUUAUUUUCGCAGGAGUUGGGCGAAUGUUUAGAGUCUAUAAUCUUCAAAUAAUGGAAAAACUUUUGCUGUUAAGCAAGAGGAAUGCCAAAGCUUAGAUUCUUU---------- .................(((((((((((...(((((..((........))..))))).(((.....((((((((....)))))))).....)))..))))).)))))).---------- ( -25.20, z-score = -0.83, R) >droVir3.scaffold_12855 5237926 114 + 10161210 -----UGUUGUUUCCCAUUGGCAUGGGCAAAUGUUUAGAGUCUAUAAUCUUCAAAUAAUGGAAAAACUUUUGCUGUUAAGCAAGAGGAAUGCCAAAGCUUAGAUUUUUUCAGAAACCAC -----.((.(((((...(((((((...(...(((((..((........))..))))).........((((((((....))))))))).)))))))......((.....)).))))).)) ( -26.00, z-score = -0.93, R) >consensus ____________UGCGGUGGGGGUGGGCGAAUGUUUAGAGUCUAUAAUCUUCAAAUAAUGGAAAAACUUUUGCUGUUAAGCAAGAGGAAUGCCAAAGCUUAGAUUUGUC__________ .....................................(((((((....(((.......(((.....((((((((....)))))))).....))))))..)))))))............. (-18.00 = -17.54 + -0.46)

| Location | 11,585,617 – 11,585,710 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 89.30 |
| Shannon entropy | 0.20943 |
| G+C content | 0.29488 |
| Mean single sequence MFE | -19.31 |
| Consensus MFE | -15.57 |
| Energy contribution | -15.35 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.587727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
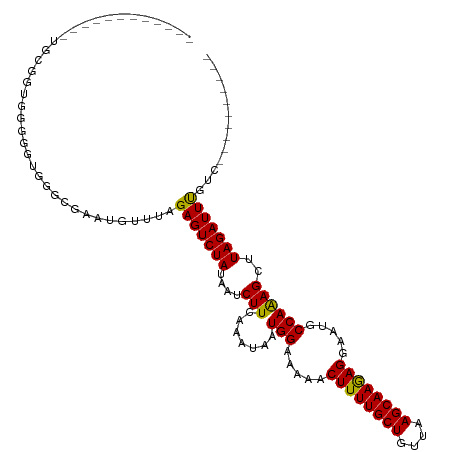
>dm3.chr3R 11585617 93 + 27905053 UCUAUAAUCUUCAAAUAAUGGAAAAACUUUUGCUGUUAAGCAAGAGGAAUGCCAAAGCUUAGAUUUGUCUUUUAUUUUGGGUUCUUCAUUUUU---------- ................((((((....((((((((....))))))))((((.((((((...(((....)))....))))))))))))))))...---------- ( -20.10, z-score = -1.54, R) >droSim1.chr3R 17617416 93 - 27517382 UCUAUAAUCUUCAAAUAAUGGAAAAACUUUUGCUGUUAAGCAAGAGGAAUGCCAAAGCUUAGAUUUGUCUUUUAUUUUGGGUUCUUCAUUUUU---------- ................((((((....((((((((....))))))))((((.((((((...(((....)))....))))))))))))))))...---------- ( -20.10, z-score = -1.54, R) >droSec1.super_0 10752111 92 - 21120651 UCUAUAAUCUUCAAAUAAUGGAAAAACUUUUGCUGUUAAGCAAGAGGAAUGCCAAAGCUUAGAUUUGUCUGUUAUUUUGGGUUCUUCUUUUU----------- (((((.((......)).)))))....((((((((....))))))))((((.((((((..((((....))))...))))))))))........----------- ( -20.10, z-score = -1.49, R) >droYak2.chr3R 13979185 103 - 28832112 UCUAUAAUCUUCAAAUAAUGGAAAAACUUUUGCUGUUAAGCAAGAGGAAUGCCAAAGCUUAGAUUUGUCUCUUAUUUUGGGUUCUUCUUUUUUUUUCAUUUUU ................((((((((((...(((((....)))))(((((((.((((((...(((......)))..)))))))))))))...))))))))))... ( -23.80, z-score = -2.48, R) >droEre2.scaffold_4770 11096483 93 + 17746568 UCUAUAAUCUUCAAAUAAUGGAAAAACUUUUGCUGUUAAGCAAGAGGAAUGCCAAAGCUUAGAUUUGUCUUUUAUUUCGGGUUCUUCAUUUUU---------- ................((((((....((((((((....))))))))((((.(.(((....(((....)))....))).).))))))))))...---------- ( -17.80, z-score = -0.91, R) >droAna3.scaffold_13340 16187507 93 + 23697760 UCUAUAAUCUUCAAAUAAUGGAAAAACUUUUGCUGUUAAGCAAAAGGAAUGCCAAAGCUUAGAUUUGUCUUUUAUUUGCUGUUCUUCAUUUUU---------- ................((((((....((((((((....))))))))((((..((((....(((....)))....))))..))))))))))...---------- ( -18.60, z-score = -1.48, R) >dp4.chr2 30629924 89 + 30794189 UCUAUAAUCUUCAAAUAAUGGAAAAACUUUUGCUGUUAAGCAAAAGGAAUGCCAGAGCUUAGAUUUGUCUUUUAUUUU---UCUACCGUUUC----------- .....(((((........(((.....((((((((....)))))))).....)))......))))).............---...........----------- ( -16.14, z-score = -1.08, R) >droPer1.super_6 6031636 89 + 6141320 UCUAUAAUCUUCAAAUAAUGGAAAAACUUUUGCUGUUAAGCAAAAGGAAUGCCAGAGCUUAGAUUUGUCUUUUAUUUU---UCUACCGUUUC----------- .....(((((........(((.....((((((((....)))))))).....)))......))))).............---...........----------- ( -16.14, z-score = -1.08, R) >droWil1.scaffold_181089 10044812 90 - 12369635 UCUAUAAUCUUCAAAUAAUGGAAAAACUUUUGCUGUUAAGCAAGAGGAAUGCCAAAGCUUAGAUUUUUCUCUG---UUGAGUUUUCCAUUUUU---------- ................(((((((((.((((((((....)))))))).........(((..(((....)))..)---))...)))))))))...---------- ( -21.00, z-score = -2.03, R) >consensus UCUAUAAUCUUCAAAUAAUGGAAAAACUUUUGCUGUUAAGCAAGAGGAAUGCCAAAGCUUAGAUUUGUCUUUUAUUUUGGGUUCUUCAUUUUU__________ .....(((((........(((.....((((((((....)))))))).....)))......)))))...................................... (-15.57 = -15.35 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:18:18 2011