| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,578,065 – 11,578,188 |
| Length | 123 |
| Max. P | 0.997779 |

| Location | 11,578,065 – 11,578,163 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 84.25 |
| Shannon entropy | 0.28878 |
| G+C content | 0.42350 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -24.80 |
| Energy contribution | -25.20 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.18 |
| SVM RNA-class probability | 0.997779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
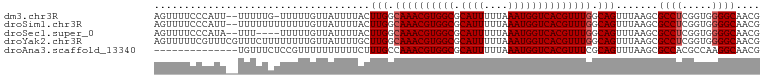
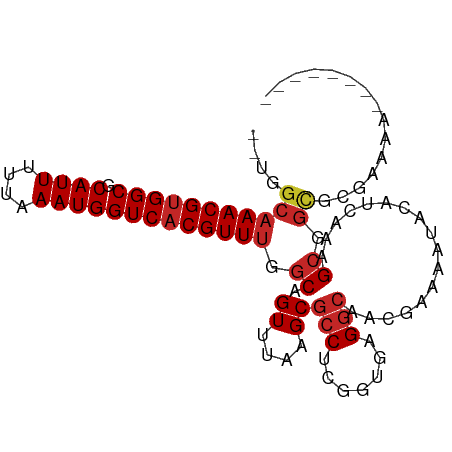
>dm3.chr3R 11578065 98 + 27905053 AGUUUUCCCAUU--UUUUUG-UUUUUGUUAUUUUACUUGGCAAACGUGGCGCAUUUUUAAAUGGUCACGUUUGGCAGUUUAAGCGCCUCGGUGGGGCAACG .(((.((((((.--......-..(((((((.......)))))))((.(((((....((((((.((((....)))).))))))))))).)))))))).))). ( -31.50, z-score = -3.40, R) >droSim1.chr3R 17609455 99 - 27517382 AGUUUUCCCAUU--UUUUUUUUUUUUGUUAUUUUACUUGGCAAACGUGGCGCAUUUUUAAAUGGUCACGUUUGGCAGUUUAAGCGCCUCGGUGGGGCAACG .(((.((((((.--.........(((((((.......)))))))((.(((((....((((((.((((....)))).))))))))))).)))))))).))). ( -31.50, z-score = -3.88, R) >droSec1.super_0 10743547 95 - 21120651 AGUUUUCCCAUA--UUU----UUUUUGUUAUUUUACUUGGCAAACGUGGCGCAUUUUUAAAUGGUCACGUUUGGCAGUUUAAGCGCCUCGGUGGGGCAACG .(((.((((((.--...----..(((((((.......)))))))((.(((((....((((((.((((....)))).))))))))))).)))))))).))). ( -32.10, z-score = -3.96, R) >droYak2.chr3R 13970652 101 - 28832112 AGUUUUUCGUUUCGUUUCUUUUUUUUGUUAUUUUGCUUGGCAAACGUGGCGCAUUUUUAAAUGGUCACGUUUGGCAGUUUAAGCGCCUCGGUGGGGCAACG ............((((..................(((((.((((((((((.((((....)))))))))))))).)).....)))((((.....)))))))) ( -29.30, z-score = -2.60, R) >droAna3.scaffold_13340 16180071 87 + 23697760 --------------UGUUUCUCCGUUUUUUUUUUCUUUGCCAAACGUGGCGCAUUUUUAAAUGGUCACGUUUCGCAGUUUAAGCGCCACGCCAAGGCAACG --------------........((((..........((((.(((((((((.((((....))))))))))))).)))).......(((.......))))))) ( -23.70, z-score = -2.14, R) >consensus AGUUUUCCCAUU__UUUUUUUUUUUUGUUAUUUUACUUGGCAAACGUGGCGCAUUUUUAAAUGGUCACGUUUGGCAGUUUAAGCGCCUCGGUGGGGCAACG ....................................(((.((((((((((.((((....)))))))))))))).))).......((((.....)))).... (-24.80 = -25.20 + 0.40)

| Location | 11,578,065 – 11,578,163 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 84.25 |
| Shannon entropy | 0.28878 |
| G+C content | 0.42350 |
| Mean single sequence MFE | -22.80 |
| Consensus MFE | -15.74 |
| Energy contribution | -15.94 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.885630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11578065 98 - 27905053 CGUUGCCCCACCGAGGCGCUUAAACUGCCAAACGUGACCAUUUAAAAAUGCGCCACGUUUGCCAAGUAAAAUAACAAAAA-CAAAAA--AAUGGGAAAACU .(((..((((....(((((.......)).(((((((.(((((....)))).).))))))))))..((......)).....-......--..))))..))). ( -22.80, z-score = -2.16, R) >droSim1.chr3R 17609455 99 + 27517382 CGUUGCCCCACCGAGGCGCUUAAACUGCCAAACGUGACCAUUUAAAAAUGCGCCACGUUUGCCAAGUAAAAUAACAAAAAAAAAAAA--AAUGGGAAAACU .(((..((((....(((((.......)).(((((((.(((((....)))).).))))))))))..((......))............--..))))..))). ( -22.80, z-score = -2.35, R) >droSec1.super_0 10743547 95 + 21120651 CGUUGCCCCACCGAGGCGCUUAAACUGCCAAACGUGACCAUUUAAAAAUGCGCCACGUUUGCCAAGUAAAAUAACAAAAA----AAA--UAUGGGAAAACU .(((..((((....(((((.......)).(((((((.(((((....)))).).))))))))))..((......)).....----...--..))))..))). ( -22.80, z-score = -2.25, R) >droYak2.chr3R 13970652 101 + 28832112 CGUUGCCCCACCGAGGCGCUUAAACUGCCAAACGUGACCAUUUAAAAAUGCGCCACGUUUGCCAAGCAAAAUAACAAAAAAAAGAAACGAAACGAAAAACU (((((((.......)))(((.....((.((((((((.(((((....)))).).)))))))).)))))..................))))............ ( -20.90, z-score = -2.43, R) >droAna3.scaffold_13340 16180071 87 - 23697760 CGUUGCCUUGGCGUGGCGCUUAAACUGCGAAACGUGACCAUUUAAAAAUGCGCCACGUUUGGCAAAGAAAAAAAAAACGGAGAAACA-------------- ..(((((..((((((((((..............................)))))))))).)))))......................-------------- ( -24.71, z-score = -1.51, R) >consensus CGUUGCCCCACCGAGGCGCUUAAACUGCCAAACGUGACCAUUUAAAAAUGCGCCACGUUUGCCAAGUAAAAUAACAAAAAAAAAAAA__AAUGGGAAAACU ....(((.......)))........((.((((((((.(((((....)))).).)))))))).))..................................... (-15.74 = -15.94 + 0.20)

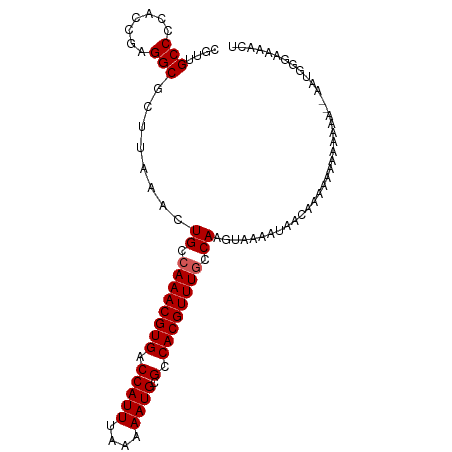
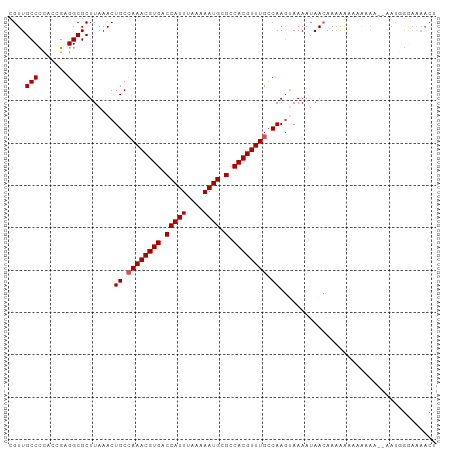
| Location | 11,578,097 – 11,578,188 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 82.57 |
| Shannon entropy | 0.34241 |
| G+C content | 0.48498 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -17.47 |
| Energy contribution | -18.09 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.855718 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 11578097 91 + 27905053 CUUGGCAAACGUGGCGCAUUUUUAAAUGGUCACGUUUGGCAGUUUAAGCGCCUCGGUGGGGCAACGAAAAUACAUCAAAGCGGCGCGAAAA-------- .(((.((((((((((.((((....)))))))))))))).))).....(((((.(..((((....)(......).)))..).))))).....-------- ( -33.00, z-score = -2.88, R) >droSim1.chr3R 17609488 91 - 27517382 CUUGGCAAACGUGGCGCAUUUUUAAAUGGUCACGUUUGGCAGUUUAAGCGCCUCGGUGGGGCAACGAAAAUACAUCAAAGCGGCGCGAAAA-------- .(((.((((((((((.((((....)))))))))))))).))).....(((((.(..((((....)(......).)))..).))))).....-------- ( -33.00, z-score = -2.88, R) >droSec1.super_0 10743576 91 - 21120651 CUUGGCAAACGUGGCGCAUUUUUAAAUGGUCACGUUUGGCAGUUUAAGCGCCUCGGUGGGGCAACGAAAAUACAUCAAAGCGGCGCGAAAA-------- .(((.((((((((((.((((....)))))))))))))).))).....(((((.(..((((....)(......).)))..).))))).....-------- ( -33.00, z-score = -2.88, R) >droEre2.scaffold_4770 11088923 91 + 17746568 CUUGGCAAACGUGGCGCAUUUUUAAAUGGUCACGUUUGGCAGUUUAAGCGCCUCGGUGGGGCAACGAAAAUACAUCAAAGCGGCGCGAAAA-------- .(((.((((((((((.((((....)))))))))))))).))).....(((((.(..((((....)(......).)))..).))))).....-------- ( -33.00, z-score = -2.88, R) >droYak2.chr3R 13970687 91 - 28832112 CUUGGCAAACGUGGCGCAUUUUUAAAUGGUCACGUUUGGCAGUUUAAGCGCCUCGGUGGGGCAACGAAAAUACAUCAAAGCGGCGCGAAAA-------- .(((.((((((((((.((((....)))))))))))))).))).....(((((.(..((((....)(......).)))..).))))).....-------- ( -33.00, z-score = -2.88, R) >droAna3.scaffold_13340 16180097 85 + 23697760 -----CAAACGUGGCGCAUUUUUAAAUGGUCACGUUUCGCAGUUUAAGCGCCACGCCAAGGCAACGAAAAUACAUCAAAGCGGCGCGAAA--------- -----.(((((((((.((((....)))))))))))))..........(((((.(((....))...((.......))...).)))))....--------- ( -26.50, z-score = -1.90, R) >droWil1.scaffold_181089 10034730 87 - 12369635 --UGGCAAACGUGGCGCAUUUUUAAAUGGUCACGUUUUUCAGUUUAAGCGCCAAAUAAAGGGAAAAGAACAACAUCAAAGCGUUGAAAA---------- --(((((((((((((.((((....)))))))))))))....((....))))))................((((........))))....---------- ( -21.50, z-score = -2.02, R) >droVir3.scaffold_12855 5227509 92 + 10161210 ---GGCAAACGUGGCGCAUUUUUAAAUGGUCACGUUUUGCAGUUUAAGCGCCAGGC--AGGCAGC--AAAUACAUCAAAGCGGCGAAAGCGACGCGAAA ---.(((((((((((.((((....)))))))))).))))).......(((((....--.))).))--............(((((....))..))).... ( -30.80, z-score = -2.02, R) >droMoj3.scaffold_6540 22500527 81 + 34148556 ---GGCAAACGUGGCGCAUUUUUAAAUGGUCACGUUCUGCAGUUUAAGCGCCUGGC--AGCCAGC--AAAUACAUCAAAGCGGCGAAA----------- ---.(((((((((((.((((....)))))))))))).)))........(((((((.--..)))((--............))))))...----------- ( -23.90, z-score = -0.64, R) >droGri2.scaffold_14906 4891492 92 - 14172833 ---GGCAAACGGGGCGCAUUUUUAAAUGGUCACGUUUUGCAGUUUAAGCGCCAGGC--AGGCAGC--AAAUACAUCAAAGCGGCGAAGGCGACGCGAAA ---.(((((((.(((.((((....))))))).)).))))).......(((((....--.))).))--............(((((....))..))).... ( -25.00, z-score = -0.27, R) >consensus __UGGCAAACGUGGCGCAUUUUUAAAUGGUCACGUUUGGCAGUUUAAGCGCCUCGGUGAGGCAACGAAAAUACAUCAAAGCGGCGCGAAAA________ ....(((((((((((.((((....))))))))))))).((.((....))(((.......))).................)).))............... (-17.47 = -18.09 + 0.62)

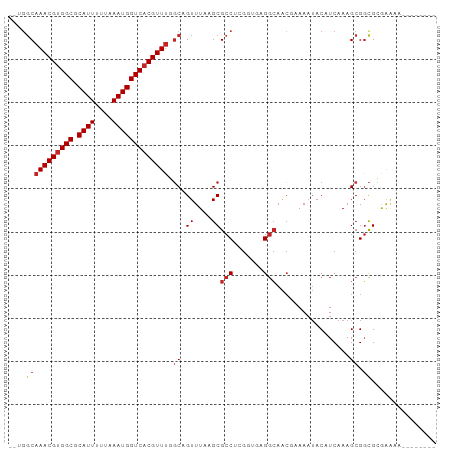
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:18:15 2011