| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,556,009 – 7,556,106 |
| Length | 97 |
| Max. P | 0.538254 |

| Location | 7,556,009 – 7,556,106 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 76.46 |
| Shannon entropy | 0.47946 |
| G+C content | 0.47748 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -11.23 |
| Energy contribution | -11.50 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.538254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
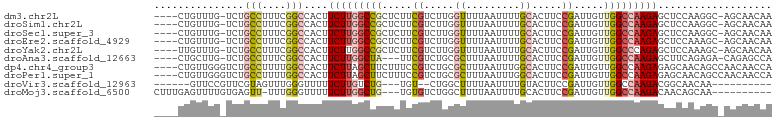
>dm3.chr2L 7556009 97 + 23011544 ----CUGUUUG-UCUGCCUUUCGGCCACUUCUUGGCCGCUCUUCGUCUUGGUUUUAAUUUUGCACUUCCGAUUGUUGGCCAAGAGCUCCAAGGC-AGCAACAA ----.((((.(-.(((((((..(((((.....)))))((((((.(((((((................)))).....))).))))))...)))))-))))))). ( -32.69, z-score = -2.97, R) >droSim1.chr2L 7351649 97 + 22036055 ----CUGUUUG-UCUGCCUUUCGGCCACUUCUUGGCCGCUCUUCGUCUUGGUUUUAAUUUUGCACUUCCGAUUGUUGGCCAAGAGCUCCAAGGC-AGCAACAA ----.((((.(-.(((((((..(((((.....)))))((((((.(((((((................)))).....))).))))))...)))))-))))))). ( -32.69, z-score = -2.97, R) >droSec1.super_3 3069692 97 + 7220098 ----CUGUUUG-UCUGCCUUUCGGCCACUUCUUGGCCGCUCUUCGUCUUGGUUUUAAUUUUGCACUUCCGAUUGUUGGCCAAGAGCUCCAAGGC-AGCAACAA ----.((((.(-.(((((((..(((((.....)))))((((((.(((((((................)))).....))).))))))...)))))-))))))). ( -32.69, z-score = -2.97, R) >droEre2.scaffold_4929 16474340 97 + 26641161 ----CUGUUUG-UCUGCCUUUCGGCCACUUCUUGGCCGCUCUUCGUCUUGGUUUUAAUUUUGCACUUCCGAUUGUUGGCCAAGAGCUCCAAAGC-AGCAACAA ----.((((.(-.((((.....(((((.....)))))((((((.(((((((................)))).....))).))))))......))-))))))). ( -27.49, z-score = -1.66, R) >droYak2.chr2L 16983533 97 - 22324452 ----UUGUUUG-UCUGCCUUUCGGCCACUUCUUGGCCGCUCUUCGUCUUGGUUUUAAUUUUGCACUUCCGAUUGUUGGCCCAGAGCUCCAAAGC-AGCAACAA ----.((((.(-.((((.....(((((.....)))))(((((..(((((((................)))).....)))..)))))......))-))))))). ( -25.59, z-score = -1.10, R) >droAna3.scaffold_12663 5834 94 + 19816 ----CUGCUUG-UCUGCCUUUCGGCCACUUCUUGGCUA---UUCGUCUGCGCUUUAAUUUUGCACUUCCGAUUGUUGGCCAAGAGCUUCAGAGA-CAGAGCCA ----..(((((-((((((....)))...((((((((((---.(((...(.((.........)).)...)))....))))))))))......)))-)).))).. ( -29.10, z-score = -1.62, R) >dp4.chr4_group3 1430856 99 + 11692001 ----CUGUUGGGUCUGCCUUUUGGCCACUUCUUAGCUUCUUUCCGUCUGCGCUUUAAUUUGGCACUUCCGAUUGUUGGCCAAGAGAGCAACAGCCAACAACCA ----.((((((.(.((((((((((((((.((...((............))(((.......)))......))..).)))))))))).)))..).)))))).... ( -31.10, z-score = -2.00, R) >droPer1.super_1 2899586 99 + 10282868 ----CUGUUGGGUCUGCCUUUUGGCCACUUCUUAGCUUCUUUCCGUCUGCGCUUUAAUUUGGCACUUCCGAUUGUUGGCCAAGAGAGCAACAGCCAACAACCA ----.((((((.(.((((((((((((((.((...((............))(((.......)))......))..).)))))))))).)))..).)))))).... ( -31.10, z-score = -2.00, R) >droVir3.scaffold_12963 9195133 82 - 20206255 ------GUUCCGUUCGUAGUUUGGGUUUUUCUUGUCUG---UGU--CUGGCUUUUAAUUUUGUACUUCCGAUUGUUGGCCAAGACGGCAACAA---------- ------(((((((((((((...(((....)))...)))---)).--.(((((..(((((..........)))))..))))).))))).)))..---------- ( -18.00, z-score = -0.85, R) >droMoj3.scaffold_6500 12607494 89 - 32352404 CUUUGAGUUUUGUGAGUU-UUUGGGUUUUUCUUGGCUG---UGUGUCUGGCUUUUAAUUUUGCACUUCCGAUUGUUGGCCAAGACAACAGCAA---------- ..................-...............((((---(.(((((((((..(((((..........)))))..)))).))))))))))..---------- ( -21.60, z-score = -0.63, R) >consensus ____CUGUUUG_UCUGCCUUUCGGCCACUUCUUGGCCGCUCUUCGUCUGGGUUUUAAUUUUGCACUUCCGAUUGUUGGCCAAGAGCUCCAAAGC_AGCAACAA ...............(((....)))....(((((((((.....((.....((.........)).....)).....)))))))))................... (-11.23 = -11.50 + 0.27)


| Location | 7,556,009 – 7,556,106 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 76.46 |
| Shannon entropy | 0.47946 |
| G+C content | 0.47748 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -11.59 |
| Energy contribution | -12.61 |
| Covariance contribution | 1.02 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2L 7556009 97 - 23011544 UUGUUGCU-GCCUUGGAGCUCUUGGCCAACAAUCGGAAGUGCAAAAUUAAAACCAAGACGAAGAGCGGCCAAGAAGUGGCCGAAAGGCAGA-CAAACAG---- .((((.((-(((((((.((((((((((......(....).((......................))))))))).)))..))..))))))).-..)))).---- ( -32.45, z-score = -2.64, R) >droSim1.chr2L 7351649 97 - 22036055 UUGUUGCU-GCCUUGGAGCUCUUGGCCAACAAUCGGAAGUGCAAAAUUAAAACCAAGACGAAGAGCGGCCAAGAAGUGGCCGAAAGGCAGA-CAAACAG---- .((((.((-(((((((.((((((((((......(....).((......................))))))))).)))..))..))))))).-..)))).---- ( -32.45, z-score = -2.64, R) >droSec1.super_3 3069692 97 - 7220098 UUGUUGCU-GCCUUGGAGCUCUUGGCCAACAAUCGGAAGUGCAAAAUUAAAACCAAGACGAAGAGCGGCCAAGAAGUGGCCGAAAGGCAGA-CAAACAG---- .((((.((-(((((((.((((((((((......(....).((......................))))))))).)))..))..))))))).-..)))).---- ( -32.45, z-score = -2.64, R) >droEre2.scaffold_4929 16474340 97 - 26641161 UUGUUGCU-GCUUUGGAGCUCUUGGCCAACAAUCGGAAGUGCAAAAUUAAAACCAAGACGAAGAGCGGCCAAGAAGUGGCCGAAAGGCAGA-CAAACAG---- .((((.((-(((((((.((((((((((......(....).((......................))))))))).)))..))))..))))).-..)))).---- ( -30.45, z-score = -2.07, R) >droYak2.chr2L 16983533 97 + 22324452 UUGUUGCU-GCUUUGGAGCUCUGGGCCAACAAUCGGAAGUGCAAAAUUAAAACCAAGACGAAGAGCGGCCAAGAAGUGGCCGAAAGGCAGA-CAAACAA---- .((((.((-(((((...((.((...((.......)).)).)).......................((((((.....)))))).))))))).-..)))).---- ( -27.50, z-score = -0.94, R) >droAna3.scaffold_12663 5834 94 - 19816 UGGCUCUG-UCUCUGAAGCUCUUGGCCAACAAUCGGAAGUGCAAAAUUAAAGCGCAGACGAA---UAGCCAAGAAGUGGCCGAAAGGCAGA-CAAGCAG---- ..(((.((-(((.....(((((((((......(((...((((.........))))...))).---..)))))).))).(((....))))))-)))))..---- ( -33.60, z-score = -2.89, R) >dp4.chr4_group3 1430856 99 - 11692001 UGGUUGUUGGCUGUUGCUCUCUUGGCCAACAAUCGGAAGUGCCAAAUUAAAGCGCAGACGGAAAGAAGCUAAGAAGUGGCCAAAAGGCAGACCCAACAG---- ....((((((...((((.((.((((((.....(((...((((.........))))...)))......(((....))))))))).))))))..)))))).---- ( -30.80, z-score = -1.24, R) >droPer1.super_1 2899586 99 - 10282868 UGGUUGUUGGCUGUUGCUCUCUUGGCCAACAAUCGGAAGUGCCAAAUUAAAGCGCAGACGGAAAGAAGCUAAGAAGUGGCCAAAAGGCAGACCCAACAG---- ....((((((...((((.((.((((((.....(((...((((.........))))...)))......(((....))))))))).))))))..)))))).---- ( -30.80, z-score = -1.24, R) >droVir3.scaffold_12963 9195133 82 + 20206255 ----------UUGUUGCCGUCUUGGCCAACAAUCGGAAGUACAAAAUUAAAAGCCAG--ACA---CAGACAAGAAAAACCCAAACUACGAACGGAAC------ ----------(((((...((((.(((.......(....).............)))))--)).---..))))).........................------ ( -10.95, z-score = 0.13, R) >droMoj3.scaffold_6500 12607494 89 + 32352404 ----------UUGCUGUUGUCUUGGCCAACAAUCGGAAGUGCAAAAUUAAAAGCCAGACACA---CAGCCAAGAAAAACCCAAA-AACUCACAAAACUCAAAG ----------..((((((((((.(((.......(....).............)))))))).)---))))...............-.................. ( -15.55, z-score = -1.22, R) >consensus UUGUUGCU_GCUUUGGAGCUCUUGGCCAACAAUCGGAAGUGCAAAAUUAAAACCAAGACGAAGAGCAGCCAAGAAGUGGCCGAAAGGCAGA_CAAACAG____ ...................(((((((......(((.......................)))......)))))))....(((....)))............... (-11.59 = -12.61 + 1.02)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:23:56 2011