
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,508,270 – 11,508,368 |
| Length | 98 |
| Max. P | 0.893066 |

| Location | 11,508,270 – 11,508,368 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 59.47 |
| Shannon entropy | 0.78622 |
| G+C content | 0.48523 |
| Mean single sequence MFE | -25.04 |
| Consensus MFE | -5.75 |
| Energy contribution | -6.08 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.893066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
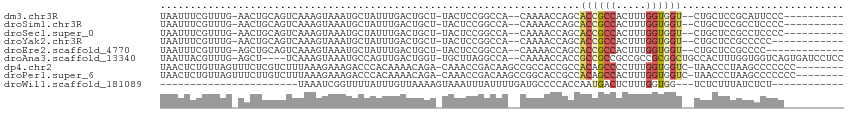
>dm3.chr3R 11508270 98 - 27905053 UAAUUUCGUUUG-AACUGCAGUCAAAGUAAAUGCUAUUUGACUGCU-UACUCCGGCCA--CAAAACCAGCACCGCCACUUUGGUGGU--CUGCUCCGCAUUCCC---------- .......((..(-(...(((((((((((....)))..)))))))).-...))..))..--......(((.((((((.....))))))--)))............---------- ( -26.30, z-score = -2.39, R) >droSim1.chr3R 17541879 98 + 27517382 UAAUUUCGUUUG-AACUGCAGUCAAAGUAAAUGCUAUUUGACUGCU-UACUCCGGCCA--CAAAACCAGCACCGCCACUUUGGUGGU--CUGCUCCGCCUCCCC---------- ............-....(((((((((((....)))..)))))))).-......(((..--......(((.((((((.....))))))--)))....))).....---------- ( -27.30, z-score = -2.88, R) >droSec1.super_0 10678822 98 + 21120651 UAAUUUCGUUUG-AACUGCAGUCAAAGUAAAUGCUAUUUGACUGCU-UACUCCGGCCA--CAAAACCAGCACCGCCACUUUGGUGGU--CUGCUCCGCCUCCCC---------- ............-....(((((((((((....)))..)))))))).-......(((..--......(((.((((((.....))))))--)))....))).....---------- ( -27.30, z-score = -2.88, R) >droYak2.chr3R 13903061 96 + 28832112 UAAUUUCGUUUG-AACUGCAGUCAAAGUAAAUGCUAUUUGACUGCU-UACUCCGGCCA--CAAAACCAGCACCGCCACUUUGGUGGU--CUGCUCCGCCCCC------------ ............-....(((((((((((....)))..)))))))).-......(((..--......(((.((((((.....))))))--)))....)))...------------ ( -26.90, z-score = -2.67, R) >droEre2.scaffold_4770 11024509 95 - 17746568 UAAUUUCGUUUG-AGCUGCAGUCAAAGUAAAUGCUAUUUGACUGCU-UACUCCGGCCA--CAAAACCAGCACCGCCACUUUGGUGGU--CUGCUCCGCCCC------------- .......((..(-((..(((((((((((....)))..)))))))).-..)))..))..--......(((.((((((.....))))))--))).........------------- ( -29.50, z-score = -2.96, R) >droAna3.scaffold_13340 16114090 106 - 23697760 UAAUUACGUUUG-AGCU----UCAAAGUAAAUGCCAGUUGACUGGU-UGCUUAGGCCA--CAAAACCACCGCCGCCGCCGCCGCGGCUGCCACUUUGGUGGUCAGUGAUCCUCC .......(((((-(((.----...........(((((....)))))-.))))))))((--(...((((((((.(((((....))))).))......))))))..)))....... ( -35.92, z-score = -1.80, R) >dp4.chr2 30516412 104 - 30794189 UAACUCUGUUAGUUUCUCGUCUUUAAAGAAAGACCCACAAAACAGA-CAAACCGACAAGCCGCCACCGCCACAGCCCCUUUGGUGGUC-UAACCCUAAGCCCCCCC-------- ......(((..((((.((((((((....))))))..........))-.))))..))).......(((((((.((...)).))))))).-.................-------- ( -18.30, z-score = -1.35, R) >droPer1.super_6 5916384 104 - 6141320 UAACUCUGUUAGUUUCUUGUCUUUAAAGAAAGACCCACAAAACAGA-CAAACCGACAAGCCGGCACCGCCACAGCCACUUUGGUGGUC-UAACCCUAAGCCCCCCC-------- ....((((((........((((((....))))))......))))))-....(((......))).(((((((.((...)).))))))).-.................-------- ( -20.84, z-score = -1.03, R) >droWil1.scaffold_181089 9950057 76 + 12369635 -----------------------UAAAUCGGUUUUAUUUGUUAAAAGUAAAUUUAUUUUGAUGCCCCACCAAUGACUCUUUGGUGG---UCUCUUUAUCUCU------------ -----------------------...(((((.(((((((.....)))))))......)))))...(((((((.......)))))))---.............------------ ( -13.00, z-score = -1.61, R) >consensus UAAUUUCGUUUG_AACUGCAGUCAAAGUAAAUGCUAUUUGACUGCU_UACUCCGGCCA__CAAAACCAGCACCGCCACUUUGGUGGU__CUGCUCCGCCCCCCC__________ ......................................................................((((((.....))))))........................... ( -5.75 = -6.08 + 0.32)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:18:08 2011