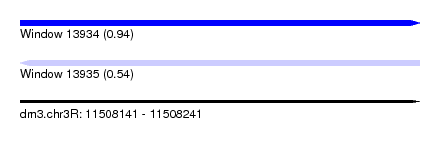
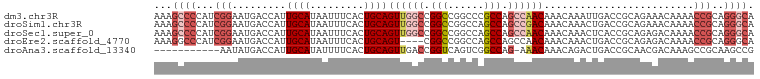
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,508,141 – 11,508,241 |
| Length | 100 |
| Max. P | 0.941660 |

| Location | 11,508,141 – 11,508,241 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 84.50 |
| Shannon entropy | 0.28186 |
| G+C content | 0.53633 |
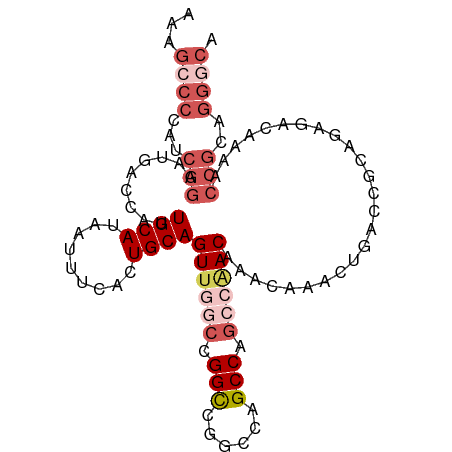
| Mean single sequence MFE | -38.70 |
| Consensus MFE | -28.14 |
| Energy contribution | -30.70 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.941660 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
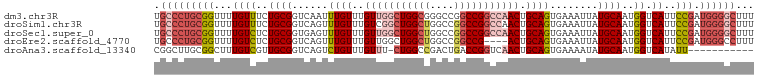
>dm3.chr3R 11508141 100 + 27905053 UGCCCUGCGGUUUUGUUUCUGCGGUCAAUUUGUUUGUUGGCUGGCGGGCCGGCCGGCCAACUGCAGUGAAAUUAUGCAAUGGUCAUUCCGAUGGGGCUUU .(((((((((...((((..((((.(((...(((..((((((((((......)))))))))).))).))).....))))..)).))..))).))))))... ( -41.40, z-score = -2.12, R) >droSim1.chr3R 17541752 100 - 27517382 UGCCCUGCGGUUUUGUUUCUGCGGUCAGUUUGUUUGUCGGCUGGCUGGCCGGCCGGCCAACUGCAGUGAAAUUAUGCAAUGGUCAUUCCGAUGGGGCUUU .(((((((((........))))(..((....(((.(((((((((....))))))))).)))((((.........)))).))..)........)))))... ( -40.40, z-score = -1.87, R) >droSec1.super_0 10678694 100 - 21120651 UGCCCUGCGGUUUUGUCUCUGCGGUGAGUUUGUUUGUUGGCUGGCUGGCCGGCCGGCCAACUGCAGUGAAAUUAUGCAAUGGUCAUUCCGAUGGGGCUUU .(((((((((...((((..((((.(((..((((..(((((((((((....))))))))))).)))).....)))))))..)).))..))).))))))... ( -43.60, z-score = -2.93, R) >droEre2.scaffold_4770 11024379 96 + 17746568 UGCCCUGCGGUUUUGUCUCUGCGGUCAGUUUGUUUGUUGGCUGGCUGGCCGGCCG----ACUGCAGUGAAAUUAUGCAAUGGUCAUUCCGAUGGGCCUUU .((((((((......((.((((((((((.....))...((((((....)))))))----))))))).)).....)))).(((.....)))..)))).... ( -38.00, z-score = -2.27, R) >droAna3.scaffold_13340 16113983 88 + 23697760 CGGCUUGCGGCUUUGUCGUUGCGGUCAGUCUGUUUGUUU-CUGGCCGACUGACCGGUCAACUGCAGUGAAAAUAUGCAAUGGUCAUAUU----------- ..((..(((((...))))).))((((((((.(((.....-..))).))))))))(..((..((((((....)).)))).))..).....----------- ( -30.10, z-score = -2.11, R) >consensus UGCCCUGCGGUUUUGUCUCUGCGGUCAGUUUGUUUGUUGGCUGGCUGGCCGGCCGGCCAACUGCAGUGAAAUUAUGCAAUGGUCAUUCCGAUGGGGCUUU .(((((((((...((((..((((......((((..(((((((((((....))))))))))).))))........))))..)).))..))).))))))... (-28.14 = -30.70 + 2.56)
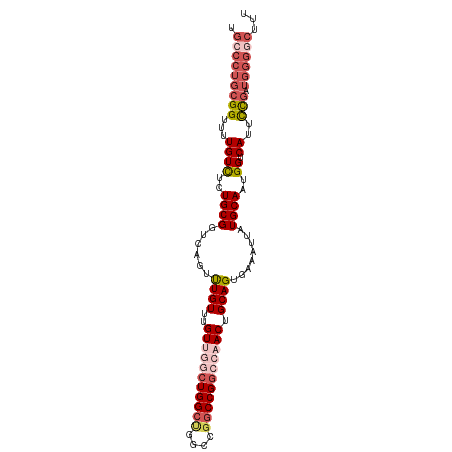
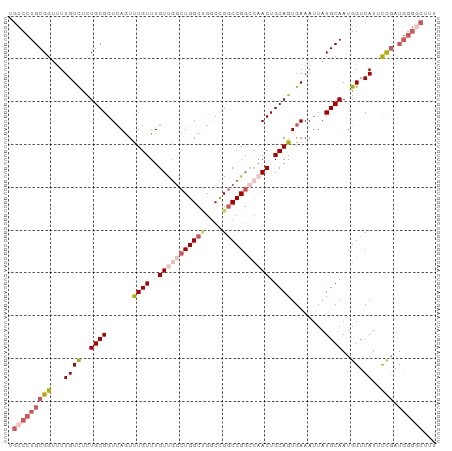
| Location | 11,508,141 – 11,508,241 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 84.50 |
| Shannon entropy | 0.28186 |
| G+C content | 0.53633 |
| Mean single sequence MFE | -29.01 |
| Consensus MFE | -17.78 |
| Energy contribution | -20.30 |
| Covariance contribution | 2.52 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.543874 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 11508141 100 - 27905053 AAAGCCCCAUCGGAAUGACCAUUGCAUAAUUUCACUGCAGUUGGCCGGCCGGCCCGCCAGCCAACAAACAAAUUGACCGCAGAAACAAAACCGCAGGGCA ...((((...(((.(((.......))).......((((.((((((.(((......))).))))))...(.....)...))))........)))..)))). ( -31.40, z-score = -1.88, R) >droSim1.chr3R 17541752 100 + 27517382 AAAGCCCCAUCGGAAUGACCAUUGCAUAAUUUCACUGCAGUUGGCCGGCCGGCCAGCCAGCCGACAAACAAACUGACCGCAGAAACAAAACCGCAGGGCA ...((((...(((.(((.......))).......((((.((((((.(((......))).))))))...(.....)...))))........)))..)))). ( -31.20, z-score = -1.82, R) >droSec1.super_0 10678694 100 + 21120651 AAAGCCCCAUCGGAAUGACCAUUGCAUAAUUUCACUGCAGUUGGCCGGCCGGCCAGCCAGCCAACAAACAAACUCACCGCAGAGACAAAACCGCAGGGCA ...((((...(((..((.....((((.........))))((((((.(((......))).))))))...))..(((......)))......)))..)))). ( -31.90, z-score = -2.39, R) >droEre2.scaffold_4770 11024379 96 - 17746568 AAAGGCCCAUCGGAAUGACCAUUGCAUAAUUUCACUGCAGU----CGGCCGGCCAGCCAGCCAACAAACAAACUGACCGCAGAGACAAAACCGCAGGGCA ....((((..(((.........((((.........))))((----((((.((....)).)))..........(((....))).)))....)))..)))). ( -28.30, z-score = -1.72, R) >droAna3.scaffold_13340 16113983 88 - 23697760 -----------AAUAUGACCAUUGCAUAUUUUCACUGCAGUUGACCGGUCAGUCGGCCAG-AAACAAACAGACUGACCGCAACGACAAAGCCGCAAGCCG -----------((((((.......)))))).....((..((((..(((((((((......-.........)))))))))))))..))..(.(....).). ( -22.26, z-score = -2.01, R) >consensus AAAGCCCCAUCGGAAUGACCAUUGCAUAAUUUCACUGCAGUUGGCCGGCCGGCCAGCCAGCCAACAAACAAACUGACCGCAGAGACAAAACCGCAGGGCA ...((((...(((.........((((.........))))((((((.(((......))).)))))).........................)))..)))). (-17.78 = -20.30 + 2.52)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:18:07 2011